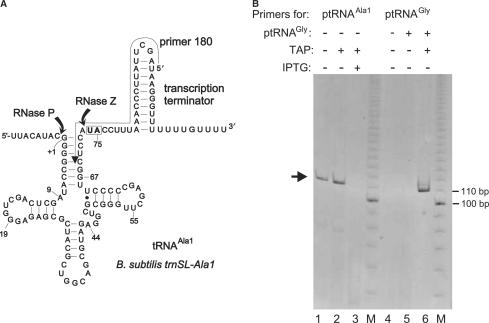
Figure 5.
5′-RACE analysis of B. subtilis trnSL-Ala1 3′-precursors. (A) Secondary structure of tRNAAla1 primary transcripts with RNase P and Z cleavage sites indicated; the arrow aligning the transcription terminator and tRNA acceptor stem indicates the position of the primer (‘180’) used for the 5′-RACE experiment; the boxed nucleotides (U74, A75) are replaced with the two C residues after RNase Z cleavage and CCA addition. (B) Analysis of 5′-RACE RT-PCR products on a native 8% PAA gel; M, 10-bp ladder as size marker; in lanes 1–3, primer ‘180’ (see above) and in lanes 4–6 a primer (see the Materials and Methods section) specific for T. thermophilus tRNAGly (Figure 1D) was used. The arrow on the left indicates the main amplification product obtained with primer ‘180’; its size was expected for a tRNAAla1 processing intermediate with a mature 5′-end but carrying the entire 3′-extension as in the primary transcript; this type of product was only detected in total RNA isolated from SSB320 cells grown in the absence of IPTG (lanes 1 and 2), but not in the presence of IPTG (lane 3); before reverse transcription, total RNA preparations were treated (+TAP) or not treated (− TAP) with tobacco acid pyrophosphatase (TAP). In lanes 5 and 6, total RNA from SSB320 cells grown in the absence of IPTG was supplemented with in vitro transcribed ptRNAGly (Figure 1D; 0.05 A260 units ptRNAGly added to 1.0 A260 unit of total cellular RNA) starting with 5′-pppG. A product corresponding to the size of ptRNAGly was only detected after TAP treatment (lane 6), but not without TAP treatment (lane 5) or without addition of in vitro transcribed tRNAGly (lane 4), thus confirming that TAP was active.