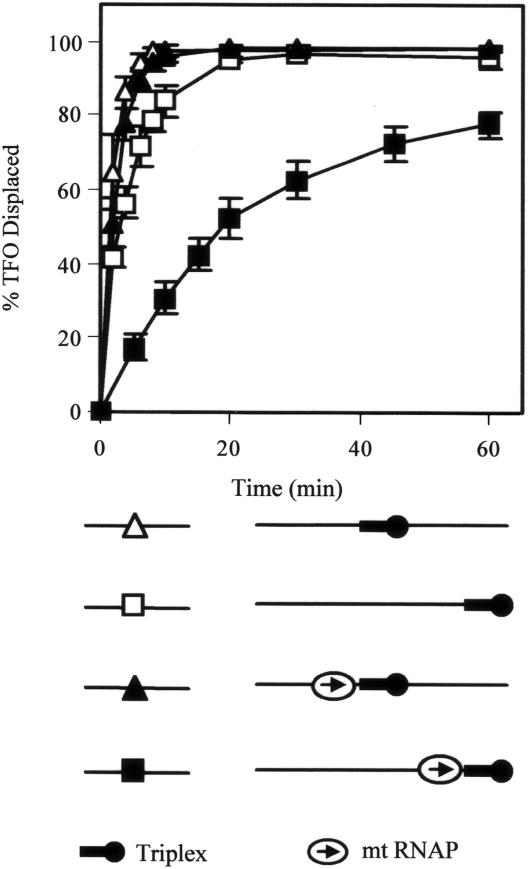
Figure 5.
The effect of flanking DNA on TFO displacement by MfdΔD7. 5 nM linear DNA fragment containing a triplex was incubated with or without 20 nM RNAP βIA117 KA118 EA119 (mt RNAP) and nucleotides (10 μM ATP, 10 μM GTP, 10 μM CTP and 100 μM ApU) for 15 min at 20°C. Reactions were initiated by the addition of 250 nM MfdΔD7 and 2 mM ATP. Aliquots were removed and the reaction quenched at the indicated time points. TFO displacement was analysed on a DNA fragment with a triplex in its middle (pSRTB1 DNA linearized with AlwNI: triangles) and a linear fragment with a triplex at its end (pSRTB1 DNA linearized with ClaI: squares). In the schematic of the substrates the 5′ end of the TFO is indicated by a black circle, and the direction in which RNAP was transcribed prior to stalling is indicated by an arrow. The graph shows the percentage of TFO displaced at each time point, normalized for the amount of triplex present at t = 0. Filled symbols indicate reactions in which elongation complexes were stalled adjacent to the triplex. Open symbols indicate reactions in which no stalled elongation complexes were present. The data are the average of at least three independent experiments, with standard deviation.