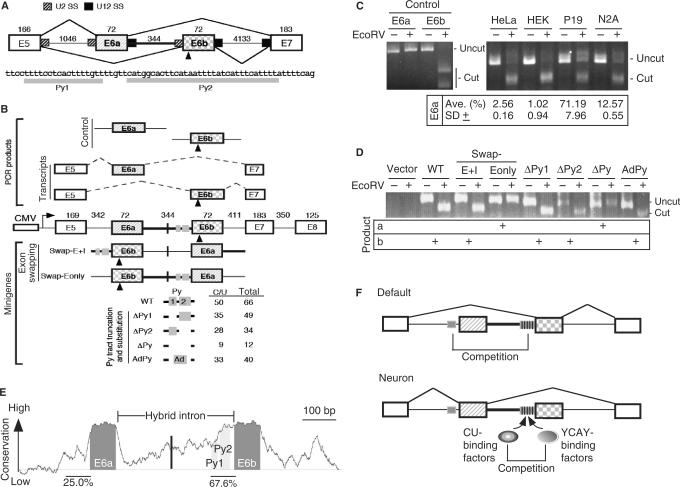
Figure 1.
The role of the U2–U12 hybrid intron in alternative splicing of human JNK2 pre-mRNA. (A) Schematic diagram shows exons 5 to 7 of JNK2. Between alternative exon 6a and 6b is the U2–U12 hybrid intron (heavy line); mutually exclusive selection of either exon generates mRNA isoforms. The numbers indicate the exon or intron length in nucleotides. U2 and U12 splice sites are indicated by hatched and filled boxes, respectively. The triangle indicates an EcoRV restriction site in exon 6b. Below the diagram is the sequence of the Py tract of the hybrid intron. (B) The minigene contains exons 5 to 8 of human JNK2, in which all introns except for the hybrid one are internally truncated (length indicated by the numbers). Transcription of the minigenes is driven by the CMV promoter. Translation start codon was in frame introduced in exon 5. Within the hybrid intron, the vertical line and grey boxes indicate the site for exon 6a/b swap and CU-rich sequences, respectively. The E6a and E6b control PCR products were amplified from the minigene, whereas the E6a and E6b transcripts represent the RT-PCR products that were amplified from the JNK transcripts. EcoRV digestion of the exon 6b-containing DNA fragment (472 bp) yielded two bands (294 and 178 bp). The modified minigenes are also depicted; the numbers of total and C/U residues of their polypyrimidine (Py) tract are listed. (C) Analysis of the JNK transcripts of four cell lines. The first four lanes show the E6a and E6b control fragments with or without EcoRV digestion. RT-PCR DNA fragments amplified from the endogenous JNK2 transcripts were of 316 bp; EcoRV digestion of the exon 6b-containing product generates two nearly comigrating fragments (cut) of 171 and 145 bp. Percentage of E6a-containing transcripts [cut/(cut + uncut) × 100%] in four cell lines was measured from three independent experiments; average with standard deviation is indicated. (D) HEK 293 cells were transfected with a minigene reporter as indicted. Total RNA was collected 24-h post-transfection and analyzed as in panel B. Shown on the gel are uncut PCR products (634 bp) and the larger digested fragment (463 bp). (E) The graph represents absolute sequence complexity (Y-axis) of a JNK2 genomic segment containing the alternative exon 6a/b and the hybrid intron (X-axis) from eight vertebrates (human, chimp, dog, cow, rat, mouse, opossum and chicken). Conservation of the Py tracts is indicated as percentage. (F) Model shows that mutually exclusive exon (hatched and squared boxes) selection of the JNK2 gene is driven by the U2–U12 intron (heavy line). The Py tract (grey box) of the hybrid intron dominates over that upstream of exon 6a, thus leading to exon 6b inclusion as the default pathway, particularly in non-neuronal cells. In neuronal cells, specific splicing activators, such as Nova, induce the use of exon 6a (21,22). On the other hand, since several YCAY elements (vertical lines) exist in the Py tract of the hybrid intron, YCAY-binding factors (such as Nova) may antagonize the activity of the ubiquitous CU-rich element-binding factors to reduce the utilization of exon 6b.