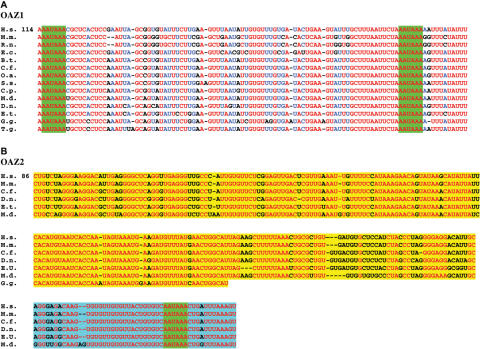
Figure 7.
Nucleotide alignment of conserved elements in the 3′ UTR of vertebrate antizyme genes. Gaps in alignment are shown by ‘-‘. The number of the top line in each case indicates the distance to the stop codon of ORF2 in human. (A) Alignment of the conserved region in orthologs of antizyme 1. Absolutely conserved nucleotides are in red and those conserved in at least 11 of the 14 species are in blue. Less well-conserved positions are in black. The two heptanucleotide sequences matching the consensus polyadenylation site are highlighted in green. (B) Absolutely conserved nucleotides are in red. Less well-conserved positions are in black. The upstream conserved region is highlighted in yellow. The downstream conserved region is highlighted in blue. The polyadenylation sites are highlighted in green. Species abbreviations are as follows: H.s. = human, M.m. = mouse, R.n. = rat, C.f. = dog, D.n. = nine-banded armadillo, E.c. = horse, B.t. = cow, O.a. = sheep, S.s. = pig, C.p. = guinea pig, E.t. = Madagascar hedgehog, M.d. = opossum, G.g. = chicken, T.g. = zebra finch.