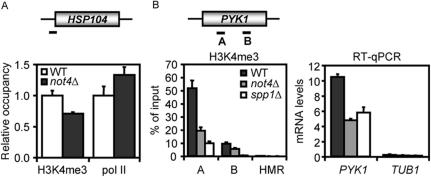
Figure 4.
Decreased H3K4me3 in not4Δ cells is not necessarily due to decreased RNA polymerase II loading (A) ChIP analysis of H3K4me3 and RNA pol II levels on the HSP104 locus. Data are represented as relative to WT. ChIP efficiencies were 2–3% and 0.15–0.2% for H3K4me3 and 8WG16 antibodies, respectively. (B) Deletion of NOT4 or SPP1 lead to a similar decrease in H3K4me3 and transcript levels on the PYK1 gene. Chromatin extracts of BY4741, not4Δ and spp1Δ strains were subjected to ChIP analysis using H3K4me3-specific antibodies. PYK1 mRNA transcript levels are affected by deletion of NOT4 or SPP1. PYK1 mRNA in total RNA of BY4147, not4Δ and spp1Δ strains was analyzed by quantitative reverse-transcriptase PCR.