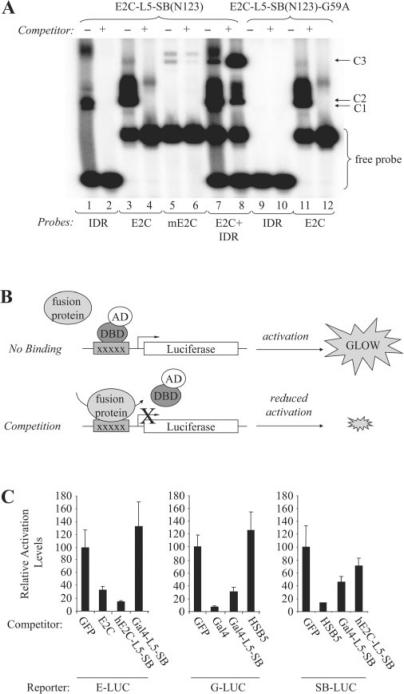
Figure 4.
DNA-binding activity of the E2C-L5-SB fusion protein. (A) Analysis of E2C-L5-SB fusion protein DNA-binding activity by electro-mobility shift assay. N123-based peptides corresponding to truncated forms of E2C-L5-SB and E2C-L5-SB-G59A (control) fusion proteins were produced by in vitro transcription and translation, and then incubated with 32P-radiolabelled double-stranded DNA probes (IDR, e2c or mutant e2c). Protein–DNA complexes were formed in the presence and absence of excess unlabelled competitor DNA, resolved by electrophoresis through a gel, and visualized by autoradiography. The competitor in lane 8 is unlabelled IDR DNA. C1, fusion peptide–IDR complex; C2, fusion peptide–e2c complex; C3, fusion peptide/IDR/e2c trimeric complex. (B) Schematic of competition assay to monitor the DNA-binding activity of full-length hE2C-L5-SB and Gal4-L5-SB fusion proteins within human cells. HeLa cells were transfected with luciferase reporter plasmids together with limiting amounts of an activator plasmid encoding their respective trans-activator protein (hE2C-AD, Gal4-AD or SB(N123)-AD). Cells also received an excess of plasmids encoding various experimental and control proteins to test whether any could compete for protein binding at the target sites, thereby reducing the level of luciferase trans-activation in the cell. (C) Reporter assay. Each graph displays luciferase activation levels relative to transfection with a control vector (pc-GFP). Bars represent the average (mean ± st.dev.) obtained from three independent transfection experiments.