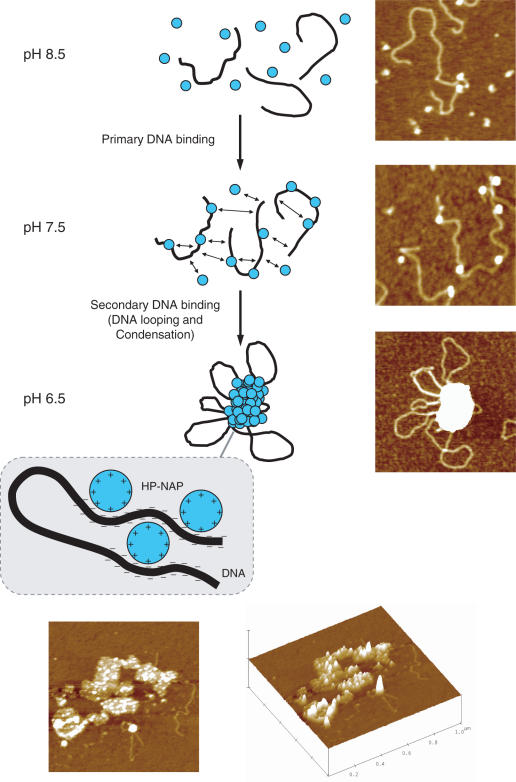
Figure 5.
Proposed model and representative images of DNA binding and condensation by H.pylori HP-NAP. At pH 8.5, HP-NAP dodecamers (blue circles) and DNA (black line) are mainly unbound. A reduction of pH to 7.5 increases the overall positive surface charge of HP-NAP which becomes able to bind DNA non-specifically by interaction with the negatively charged phosphate groups. This primary DNA-binding event generates complexes with a ‘beads-on-a-string’ morphology. Further reduction of pH to 6.5 causes the protonation of surface-exposed amino acid residues (likely histidine residues). Thereby intra- and inter-strand DNA-binding of HP-NAP is rendered possible. This secondary DNA-binding event results in DNA looping and condensation. Thus, HP-NAP–DNA condensation relies on full protonation of amino acid residues which render the overall protein surface positive and capable of multiple binding to the negatively charged DNA polymer. The top and lateral 3D view of selected HP-NAP–DNA condensates at pH 7.0 is given at the bottom. The image scan size is 1 μm.