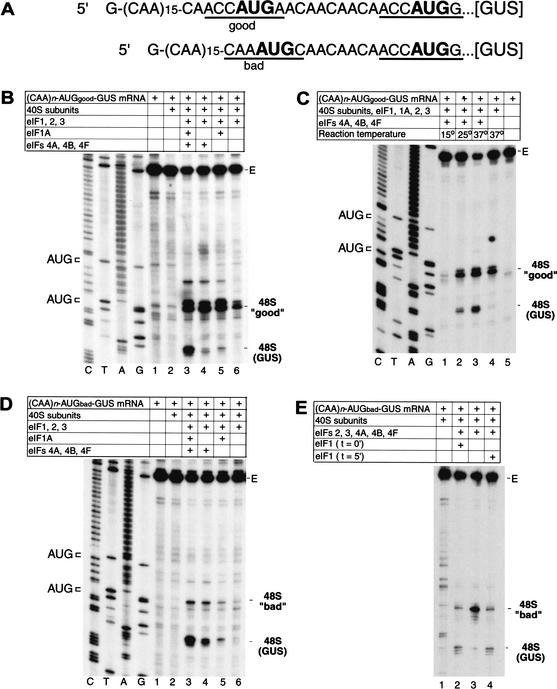
Figure 5.
Factor dependence of initiation codon selection on mRNAs containing tandem initiation codons differing in sequence context. (A) Sequences of the 5′-UTRs of (CAA)n-AUGgood-GUS and (CAA)n-AUGbad-GUS mRNAs, showing the AUG initiation codons in bold. Context residues from −3 to +4 positions for each initiation codon are underlined; the A of the AUG codon is designated as +1. (B–E) Toeprinting analysis of 48S complex formation on (CAA)n-AUGgood-GUS and (CAA)n-AUGbad-GUS mRNAs, as indicated, in reaction mixtures containing 40S subunits, GMP-PNP, ATP, aminoacylated total tRNA, and translation components as indicated and incubated at 37°C (B,D,E) or at temperatures as indicated (C). eIF1 was included at the beginning of each assembly reaction except where indicated in panel E. Full-length cDNA is labeled E in panels B–E. The label “48S (GUS)” indicates the position of toeprints caused by 48S complexes assembled on the GUS initiation codon; the labels 48S “good” and 48S “bad” indicate 48 complexes assembled at upstream initiation codons in these mRNAs that are in either good or bad context. The position of these initiation codons are shown to the left of appropriate reference lanes (C,T,A,G) in panels B–D, which show cDNA sequences derived using the same primer as for toeprinting.