Figure 5.
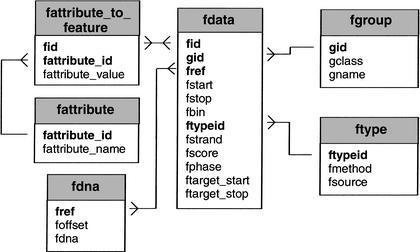
The Bio::DB::GFF database uses a minimal schema to represent features on sequences. The main tables are fdata, which contains the position and type of each feature, fgroup, which tracks the grouping of subfeatures into features, such as high-similarity pairs in a gapped alignment, fdna, which stores the raw DNA sequence, and fattribute_to_feature, which allows attribute information to be attached to features. Attributes are used for storing such textual information as notes, synonyms, and evidence codes. The fattribute and ftype tables, respectively, hold attribute names and the method and source fields. For retrieval efficiency, the fdna table fragments each DNA into small pieces and stores the beginning of each piece in the foffset field.