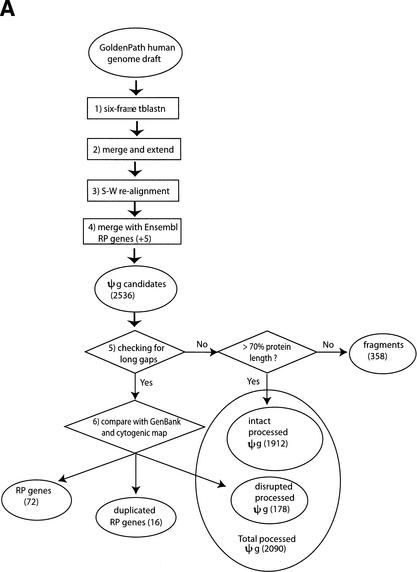
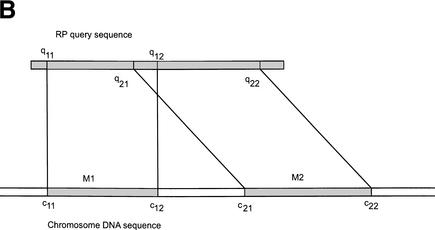
Figure 8.
(A) Flow chart of the procedure for searching for RP pseudogenes in the human genome. RP and ΨG denote “ribosomal protein” and “pseudogene”, respectively. S-W., “Smith-Waterman”. The steps are as follows: (1) Six-frame BLAST run searching for RP homologies in the human genome. (2) Merging and extension. BLAST hits were merged and extended on both sides to match the length of RP peptide sequence. (3) Smith-Waterman realignment. Extended homologies were realigned with RP sequence. (4) Comparison with Ensembl annotation. Five RPL41 pseudogenes from Ensembl were added to the set. A total of 2536 PR genes or pseudogenes were identified. (5) Checking for long gaps. Homology sequences that contained gaps shorter than 60 bp were labeled “intact processed pseudogenes” if they were longer than 70% of the full-length RP sequence; otherwise they were labeled “pseudogenic fragments”. (6) Comparison with GenBank and cytogenic mappings. For those RP homologies that contained long gaps (>60 bp), their sequences were compared with the RP exon structure from GenBank and their chromosomal locations were checked with cytogenic mapping. The homology sequences were assigned as functional RP genes, duplicated RP genes, and “disrupted processed pseudogenes.” The latter were processed pseudogenes whose sequences were interrupted by retrotransposons. (B) Schematic graph describing the considerations in merging two adjacent RP matches, M1 and M2. (c11, c12) and (c21, c22) are chromosomal coordinates for M1 and M2. (q11, q12) and (q21, q22) are corresponding regions on the query RP protein that they match.