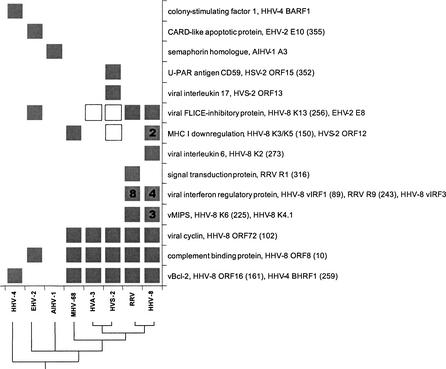
Figure 3.
Gammaherpesvirus-specific proteins involved in host-virus interactions that have human homologs. Boxes indicate the presence of a particular gene(s) in a virus genome. Numbers in boxes represent copies within a genome. Labels show the virus protein function, the name of a member of the HPF (homologous protein family) or singleton, and, for HPFs, the corresponding number in brackets. All the annotations and HPF numbers are taken from VIDA. Note that in some cases more than one HPF/singleton, shown as separate rows in Table 1, is shown together. This corresponds to highly divergent families. The HPF/singletons that are not present in Table 1 are represented as unfilled boxes. These are herpesvirus proteins for which we did not identify human homologs in the database searches but that, nevertheless, can be grouped together, by function and residue conservation, with other herpesvirus HPF/singletons for which we could detect human homologs. A consensus phylogenetic tree of the gammaherpesvirus is shown at the bottom. This was generated as described for all HPFs from complete herpesvirus genomes (Albà et al. 2001a).