Figure 1.
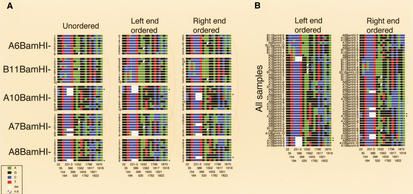
Variant box. For haplotype analysis, the combinations of a set of 18 “common” variants were diagramed for each of the individual 2-kb BamHI subclones from the 5 PAC samples. Little squares were drawn for each nucleotide using the ABI color code (see inset). Variant positions (bottom) belong to the numbering of the satellite sequence in this work (see Supplementary Fig. 1; available online at http://www.genome.org). The 3-bp deletion variant at position 231–233 spans three squares. The variants initially were isolated because they were abundant within a single X-chromosome array and typically revealed a fixed transition state within the population. Within the most homogeneous PACs A7, A8, and A10, such common variants account for the majority of the total variation. The slightly lower homogeneity of PACs A6 and B11 is mainly attributable to a higher fraction of regional variants, possibly owing to a reduced participation in the overall homogenization process (spatially or temporally). The overall sequence divergence of PACs A6 and B11 could well be considerably higher, because a significant fraction of dispersed BamHI fragments with sizes other than 2 kb were not analyzed (Table 2). Of the 3 most homogeneous PACs, only A8 contains such a disruption of the regular BamHI higher-order repeat structure (data not shown). Ordering the combinations of the 18 common variants derived from PAC samples (A) or from all PACs (B) along their left or right ends, neither led to regular patterns reminiscent of a limited number of major haplotypes, nor revealed any obvious relatedness of haplotypes with respect to PAC origin. Of the 46 combinations from individual subclones, 40 are unique and none occurs three times. The six pairs of combinations occurring twice were found within the highly homogeneous PAC A7 (2×), the less homogeneous PAC B11 (containing a duplication of 35–50 kb), between the two homogeneous PACs A10 and A8, or between the homogeneous PAC A7 and diverged PAC A6, as indicated by arches. Asterisks indicate individual clones with identical end sequences. Several combinations of PAC B11 seem to be more related to each other than to the combinations of other PACs, which to some extent could be explained by a somewhat reduced information from the set of 18 variants in this PAC. Interestingly, according to partial restriction mapping, PACs B11, A6, and A7 cannot extensively overlap with each other, and all three cannot extensively overlap with PACs A8 and A10 (which could overlap almost entirely). Nevertheless, ordering the combinations from all PACs results in many possible relationships within and between the PACs. This analysis indicates a lack of major haplotypes within the array, not supporting homogenization via amplification and replacement. In addition, similar haplotype analyses within narrow regions derived from the two or three DraIII and non-DraIII clusters of PAC A7 (see Supplementary Figs. 3, 5; available online at http://www.genome.org) indicate that small regions of amplified haplotypes are also very unlikely.