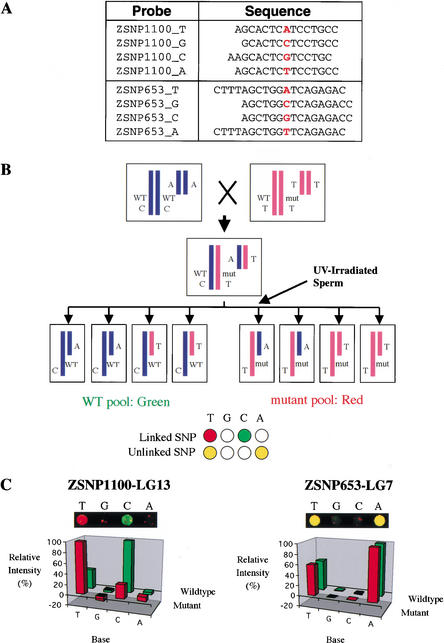
Figure 3.
(A) Oligonucleotide probes specific for two SNPs. (B) Mapping scheme with haploid crosses. Haploid embryos are generated from F1 females heterozygous for the mutation and for many SNPs (two of which are shown). SNPs are scored in pooled wild-type and mutant samples by hybridizing differentially labeled fragments to the same microarray. Alleles of linked SNPs are differentially labeled, whereas unlinked SNPs have both labels on each allele. Putative locations indicated by the microarray analysis are then tested by scoring markers in the region in individuals (not shown). (C) Microarray data mapping the floating head mutation to LG13. A two-color image from the hybridized microarray and a graph of relative hybridization intensity for the four probes of two SNPs are shown. ZSNP1100 on LG 13 exhibited differential labeling characteristic of linked markers. Polymorphic markers at other positions, including ZSNP653 on LG7, showed labeling characteristic of unlinked markers. In another trial of this experiment, we obtained similar results: Differential labeling indicative of linkage was detected for ZSNP1100, and there were no false positive or false negative markers (data not shown).