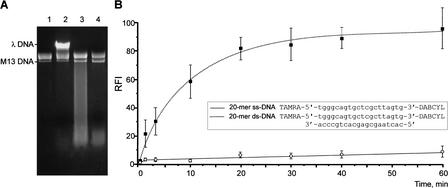
Figure 1.
Determination of crab nuclease preference for specific structural features of DNA substrates. (A) Action of crab nuclease on ss phage M13 DNA and ds λ DNA. Lanes 1,2: negative controls, incubation without nuclease. Lane 1: phage M13 DNA alone; lane 2, mixture containing phage M13 DNA and λ DNA. Lanes 3,4: Digestion of phage M13 and λ DNA mixture by crab nuclease at 70°C for 1.5 min (lane 3) and 5 min (lane 4). For experimental details, see Methods. (B) Action of crab nuclease on synthetic ss and ds 20-mer DNA substrates. The cleavage reaction was performed as described in Methods at 35°C for different periods. Fluorescence intensity was measured at 570 nm (with excitation at 550 nm). The relative fluorescence increase in the oligonucleotide substrate, RFI, was defined as RFI = (Fi − Fo/Fmax − Fo) × 100%, where Fi is the fluorescence intensity of a substrate after incubation with nuclease, Fo is the substrate fluorescence in the absence of enzyme, and Fmax represents the fluorescence of 100% cleaved substrate. For kinetic graph construction, three identical experiments were performed, and the average values and standard deviations were plotted.