Abstract
Ever since the advent of molecular methods, the diagnostics of Neisseria gonorrhoeae has been troubled by false negative and false positive results compared with culture. Commensal Neisseria species and Neisseria meningitidis are closely related to N. gonorrhoeae and may cross-react when using molecular tests comprising too-low specificity. We have devised a real-time polymerase chain reaction (PCR), including an internal amplification control, that targets the N. gonorrhoeae porA pseudogene. DNA was automatically isolated on a BioRobot M48. Our subsequent PCR method amplified all of the different N. gonorrhoeae international reference strains (n = 34) and N. gonorrhoeae clinical isolates (n = 176) but not isolates of the 13 different nongonococcal Neisseria species (n = 68) that we tested. Furthermore, a panel of gram-negative bacterial (n = 18), gram-positive bacterial (n = 23), fungal (n = 1), and viral (n = 4) as well as human DNA did not amplify. The limit of detection was determined to be less than 7.5 genome equivalents/PCR reaction. In conclusion, the N. gonorrhoeae porA pseudogene real-time PCR developed in the present study is highly sensitive, specific, robust, rapid and reproducible, making it suitable for diagnosis of N. gonorrhoeae infection.
Neisseria gonorrhoeae is a difficult organism to diagnose by culture, mainly due to its low viability in transportation media.1 This means a significant risk of false negative samples. The sensitivity of culturing may be quite low even under optimal conditions.2,3 N. gonorrhoeae detection based on techniques that are independent of bacterial viability would improve gonorrhea diagnostics radically.
N. gonorrhoeae belongs to the Neisseria genus of bacteria and is the causative agent of gonorrhea,4 which is a major sexually transmitted infection in many countries.5 The Neisseria genus comprises many closely related species,4 of which only N. meningitidis and N. gonorrhoeae are primarily pathogenic to humans. Sites of infection for N. gonorrhoeae are mainly urogenital, anorectal, and oropharyngeal mucous membranes. Many of the other Neisseria species are commensal to humans and can commonly be found on mainly extra genital mucous membranes. Neisseria species are also found on several animal hosts.4 Neisseria genomes are highly homologous,6,7,8 and interspecies genetic exchange has been described. This could be facilitated by a system secreting chromosomal DNA found in N. gonorrhoeae9 and specific uptake systems for extracellular DNA such as the one described for N. meningitidis.10
The gold standard for diagnosing N. gonorrhoeae remains culture; however, the very fastidious N. gonorrhoeae does not survive very long outside the host. This emphasizes the importance of optimized sample collection, transportation, and storage of the specimens as well as adequate culture methods. Consequently, an increasing number of nucleic acid amplification tests (NAATs) have been developed for diagnosis of N. gonorrhoeae. Commercially available NAATs have been important supplements to the laboratory diagnosis of N. gonorrhoeae, largely due to the increased sensitivity compared with culture techniques and the possibility for using noninvasive samples such as urine. However, due to the interspecies homology, concerns have frequently been raised about the specificity of many published NAATs, both commercial and in-house methods.11,12,13,14,15,16,17,18,19,20,21,22 When using NAATs for diagnosis of N. gonorrhoeae, a very high specificity is essential, especially for extragenital specimens.19 For example, the pharynx commonly harbors commensal Neisseria species and/or Neisseria meningitidis and hence is not suitable as a collection site unless a highly specific test is used.19 Some commercial NAATs have even been withdrawn from the market due to cross-reactivity with commensal Neisseria spp. and/or an insufficient analytical sensitivity.23 Due to the social impact of false positive test results, retesting of all positives with a second NAAT targeting another gene is recommended.24 This is an unacceptable solution for most laboratories as it is laborious, expensive, and time-consuming. Therefore, a highly specific NAAT that can be used for both routine analysis as well as confirmative retesting of positive samples is highly needed.
The choice of target gene is the main key to a successful N. gonorrhoeae NAAT. Previous methods have used targets such as the cryptic plasmid (cppB gene),25,26,27 opa genes,11 orf1 gene,28 cytosine DNA methyltransferase gene,18 and 16S rRNA gene.29,30 However, the cppB gene is missing in many N. gonorrhoeae strains,20,29 and most of the other targets have been shown to cross-react with the equivalent genes in commensal Neisseria species. A novel in-house PCR reported by Whiley et al31,32 that targets the porA pseudogene of N. gonorrhoeae has so far proved to have satisfactory specificity. Extensive sequencing of the porA pseudogene33,34 has supported this gene as a suitable target for N. gonorrhoeae PCR. The porA gene/pseudogene is absent in commensal Neisseria species,33,35,36 and the porA gene of N. meningitidis is sufficiently divergent to be discriminatory between the two human pathogenic Neisseria.34 Thus, this pseudogene is a nonexpressed equivalent of the N. meningitidis porA gene.33,34 There are a few inactivating mutations that make the N. gonorrhoeae porA a pseudogene.33,34 The pseudogene seems very stable, and it is not subjected to any positive selective pressure; consequently, the pseudogene has stayed virtually unchanged over time.34
Real-time PCR have made PCR more user-friendly and reduced hands-on time, time for receiving results, and level of contamination. There are several real-time PCR systems on the market that ramp temperature up to 50% faster than a conventional PCR machine. This allows the user to run up to 96 or 384 samples (40 cycles) in 35 or 55 minutes, respectively, providing a 20-μl reaction mix. These protocols are typically referred to as rapid cycle PCR or fast cycle PCR, but we call it fast real-time PCR in the present article.
In the present study, we designed and evaluated a real-time PCR for specific detection of N. gonorrhoeae that can supplement our culture method and/or be used as single diagnostic method in other geographic areas, in which adequate culturing is not possible to perform. The main priority was optimal specificity and sensitivity. We collected international N. gonorrhoeae reference strains and clinical isolates to verify the sensitivity of the method. As many isolates and international reference strains as possible of commensal Neisseria and N. meningitidis, along with other microorganisms often found in the genital area, were gathered to ensure maximum specificity. Another important criterion was that the method should be possible to automate, and an appropriate sample collection and an optimal transportation buffer should be found.
Materials and Methods
Bacterial Isolates, Clinical Samples, and Culture Conditions
International Neisseria reference strains were collected from American Type Culture Collection (ATCC), Culture Collection University of Gothenburg, National Collection of Type Cultures, World Health Organization (World Health Organization), Swedish Reference Laboratory for Pathogenic Neisseria, and Statens Serum Institut, Denmark. These reference strains included N. gonorrhoeae (n = 34), N. meningitidis (n = 4), Neisseria sicca (n = 2), Neisseria subflava (n = 1), Neisseria flavescens (n = 2), Neisseria mucosa (n = 2), Neisseria lactamica (n = 2), and Neisseria cinerea (n = 1). The N. gonorrhoeae reference strains originated from different geographic locals worldwide and have been isolated during the last four decades. Furthermore, in 29 of the 34 N. gonorrhoeae reference strains, the entire porA pseudogene has previously been sequenced.34
In addition to international reference strains, we examined 176 clinical N. gonorrhoeae isolates. These included 76 isolates cultured in Archangelsk, Russia, in 2004. These isolates were initially cultured on nonselective media (NPO Microgen, Stavropol, Russia), but species were subsequently confirmed as N. gonorrhoeae by colony characteristics on selective media,37 rapid positive oxidase test, presence of gram-negative diplococci in microscopy, sugar oxidation test,38 and specific monoclonal antibodies (Phadebact GC Monoclonal test; Boule Diagnostics AB, Huddinge, Sweden). Fourteen isolates were cultured at University Hospital of North Norway, 2003 to 2004, and nine isolates were received from Norwegian Organization for Surveillance of Antibiotic Resistant Microorganisms; all these were identified as N. gonorrhoeae by rapid positive oxidase test, the presence of gram-negative diplococci, sugar oxidation test,38 and specific monoclonal antibodies (Phadebact GC Monoclonal test). Thirteen isolates from Statens Serum Institut in Denmark were species identified by culturing on selective medium and MINIBACT-N.39 In addition, five confirmed N. gonorrhoeae isolates donated by Helen Palmer19 and 51 genetically distinct, Swedish-confirmed N. gonorrhoeae isolates from 1998 to 2001 with known porA pseudogene sequence34 were included.
Furthermore, clinical isolates (n = 54) of other Neisseria species were included. These comprised N. gonorrhoeae subspecies kochii (n = 4), N. meningitidis (n = 7), N. sicca (n = 7), N. subflava (n = 11), N. flavescens (n = 3), N. mucosa (n = 5), N. lactamica (n = 7), N. cinerea (n = 7), Neisseria caviae (n = 1), Neisseria animalis (n = 1), and Neisseria polysaccharea (n = 1). The commensal Neisseria were identified by sugar oxidation,38 oxidase testing, and, if needed, API NH or RapID NH (bioMerieux, La Balme-les-Grottes, France).
To exclude cross-reaction with genomes from other bacterial species, ie, non-Neisseria species, we tested DNA from a panel of gram-negative bacterial (n = 18), gram-positive bacterial (n = 23), fungal (n = 1), and viral (n = 4) as well as human DNA.
All Neisseria spp. were cultured from frozen stocks (−70°C) for 24 to 48 hours at 37°C in 5% CO2 on nonselective chocolate agar40 containing GC-Agar Base Medium CM367B (Oxoid, Basingstoke, UK), distilled water, 5% defibrinated horse blood, 25% glucose solution, and Vitox solution (Oxoid SR090H and SR090A).
Isolation of Bacterial DNA
Genomic DNA from the bacteria was isolated with a BioRobot M48 from Qiagen (Hilden, Germany) using the MagAttract DNA tissue kit with the Infectious disease protocol and an elution volume set to 100 μl, according to the manufacturer’s specifications. All bacteria and fungi were initially suspended in 200 μl of TE buffer, pH 8 (Ambion, Austin, TX), to approximately 1.5 × 108 colony-forming units (CFU)/ml before isolation of DNA. All viruses were suspended in a virus in-house transport medium made from minimum essential medium (Gibco, Carlsbad, CA), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid buffer (Gibco), and Gentamicin (Gibco).
Primer Design and PCR Conditions
Primers and probes against the N. gonorrhoeae porA pseudogene were designed as a TaqMan assay by using Primer Xpress 2.0 (Applied Biosystems, Foster City, CA) according to manufacturer’s guidelines. The forward and reverse primer sequences for the N. gonorrhoeae porA pseudogene were 5′-CCGGAACTGGTTTCATCTGATT-3′ and 5′-GTTTCAGCGGCAGCATTCA-3′, respectively. The sequence for the TaqMan probe for the porA pseudogene was 5′-FAM-CGTGAAAGTAGCAGGCGTATAGGCGGACTT-BHQ-1-3′. The primer sequences, probe sequence, and the amplicon were compared with the porA pseudogene sequences from a previous study34 as well as all of the genetic sequences deposited in GenBank.
For the N. gonorrhoeae-specific PCR, we used TaqMan Fast Universal PCR Master Mix (×2), No AmpErase UNG on a 7900 HT Fast Real-Time PCR System (Applied Biosystems). The Fast-PCR was run with 20-second activation of the polymerase at 95°C, followed by 50 cycles of 95°C for 1 second, and 60°C for 20 seconds. We used a 11.5-μl template added to 13.5 μl of reaction solution for each PCR sample. We also tested the robustness of the method by varying the sample volume in the PCR between 9 and 15 μl while keeping the reaction solution constant at 13.5 μl. The primers amplify a 102-bp fragment of the porA pseudogene. The porA primers and probe were optimized as recommended by Applied Biosystems, and a final concentration of 900 nmol/L of each primer and 200 nmol/L probe was found to be optimal.
A serial dilution of commercially available quantitated DNA from Tebu-bio (Le Perray en Yvelines, France) was used to generate a standard curve for efficiency (E) calculation as well as deciding the limit of detection. Efficiency was calculated using the formula: E = 10−1/slope − 1.41 To estimate the efficacy also of the transportation buffer and DNA preparation method used in the present study, a suspension was made of N. gonorrhoeae reference strain ATCC 19424 in phosphate-buffered saline. This was then diluted 10−1 through 10−7, and 100 μl of each dilution was plated on a modified Thayer-Martin medium40 and incubated overnight at 37°C in the presence of 5% CO2. Another 100 μl of each dilution 10−1 through 10−5 was transferred to 3 ml of UTM-RT sample transportation buffer (Copan, Brescia, Italy) and stored overnight at 4°C. The following day, 200 μl of each UTM-RT dilution was used to prepare DNA on the BioRobot M48, as previously described.
Internal Amplification Control
For inhibition control, an internal amplification control (IAC) was constructed by using composite primers that have N. gonorrhoeae specific 5′-end and a pGEM-luc plasmid (Promega, Madison, WI) specific 3′-end, which amplifies a 181-bp fragment of the β-lactamase coding region (positions 2988 to 3169). The forward and reverse primer sequences for making the IAC were 5′-GTTTCAGCGGCAGCATTCATGGTCTGACAGTTACCAATGCTTAA-3′ and 5′-CCGGAACTGGTTTCATCTGATTGGCTGGCTGGTTTATTGCTG-3′, respectively. The sequence for the TaqMan probe for the porA pseudogene was 5′-FAM-CGTGAAAGTAGCAGGCGTATAGGCGGACTT-BHQ-1-3′. This makes a DNA amplicon that is 255 bp long with ends that is recognized by the forward and reverse porA primers. Five μl of a 10−4 dilution of the pGEM-luc plasmid was used as a template in a total volume of 50-μl reaction solution with no UNG. Fifty nM of each of the composite primers were used. First, the reaction mixture was heated at 95°C for 10 minutes, then 40 cycles of 95°C for 15 seconds, and 60°C for 1 minute followed.
Results
All N. gonorrhoeae international reference strains (n = 34) and clinical N. gonorrhoeae isolates (n = 176) examined in the present study were positive in our developed in-house porA pseudogene real-time PCR. In addition, all international reference strains and clinical isolates of other Neisseria species than N. gonorrhoeae were negative. Furthermore, neither of the 46 non-Neisseria bacterial species nor the nonbacterial agents tested positive. Every test we performed to optimize the method, determine specificity, etc, or analyze samples was tested twice in parallel. In addition, the PCRs of representative strains/isolates were repeated to analyze the reproducibility and all of the initial results were confirmed.
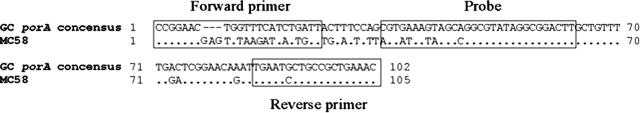
An alignment of the primers and probe on a partial consensus sequence based on recent sequencing34 of N. gonorrhoeae porA pseudogenes and the corresponding region of the N. meningitidis MC58 gene42 shows that the primer and probe regions of the N. gonorrhoeae porA pseudogene (Figure 1) is 100% conserved. Furthermore, the figure shows that the N. gonorrhoeae porA pseudogene is sufficiently different from the N. meningitidis porA gene to effectively discriminate between the two pathogenic Neisseria.
Figure 1.
Nucleotide sequence alignment of partial N. gonorrhoeae consensus porA pseudogene compiled from 87 sequences34 and the corresponding N. meningitidis sequence. The boxed sequences on the flanks are the forward and reverse primer location, respectively. The boxed sequence in the middle is the location of the probe. The MC58 sequence is the partial porA sequence of the previously published whole-genome sequenced N. meningitidis strain MC58.42
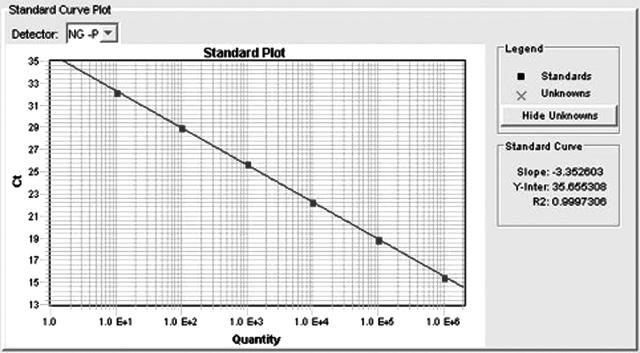
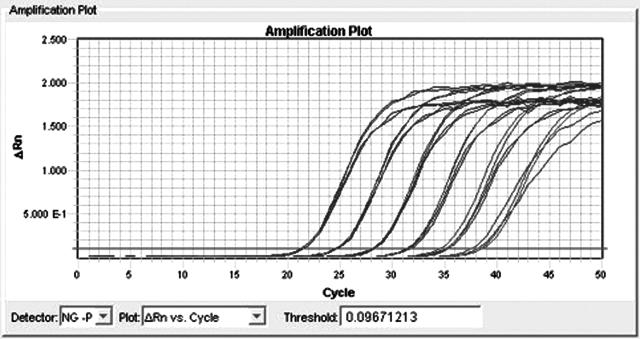
As previously mentioned, optimal concentration of each primer was shown to be 900 nmol/L. The probe concentration was optimized to 200 nmol/L of the porA probe, and 100 nmol/L of the IAC probe. The IAC was optimized to give a cycle threshold (Ct) of 37 to 39 in known negative samples. With this master mix, we see a PCR efficiency of 99% as derived from the dilution series in Figure 2 by making a standard curve (Figure 3) and using the slope in the equation E = 10−1/slope − 1. The DNA isolation procedure used in the present study can be performed in approximately 2.5 hours, whereas the present PCR takes about 40 minutes. Consequently, for 44 clinical samples (DNA isolation of two batches of samples) the total assay time is only 4 hours and 10 minutes (96 PCR reactions including controls).
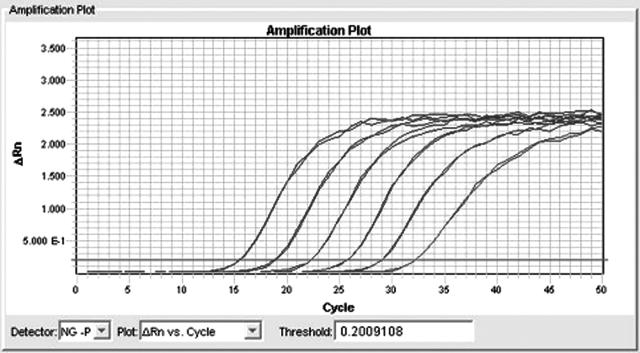
Figure 2.
Dilution series of commercially purified N. gonorrhoeae DNA (Tebu-bio) and detection in the N. gonorrhoeae porA pseudogene real-time PCR developed in the present study. The six 10-fold dilutions showed good linearity.
Figure 3.
Standard curve based on the dilution series in Figure 2. The slope is the basis for the amplification efficacy (E) calculation. In a 100% efficient PCR, the DNA will double per cycle or use 3.333 cycles to increase DNA copies by 10-fold (23.33 ≈ 10).
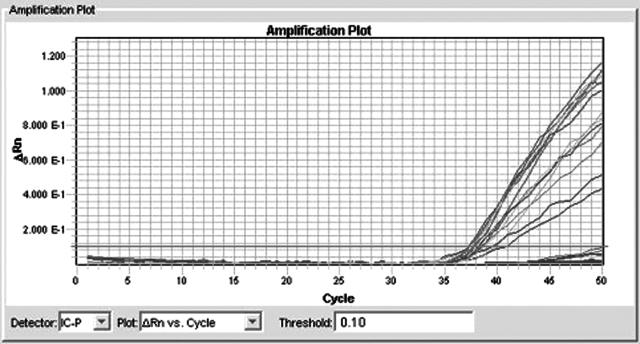
The IAC was only amplified in very low concentrations of competing N. gonorrhoeae DNA or in the total absence of N. gonorrhoeae DNA (Figure 4). A twice-repeated dilution series of N. gonorrhoeae DNA using master mixes with and without IAC showed that there was no significant competitive inhibition of the target PCR (Figure 5). The amplification efficacy was shown to be 99.3 and 94.9% for the master mix with and without IAC, respectively. The master mix with IAC had the greatest amplification efficacy. In Figure 5, the two amplification plots overlay each other with very little difference, yet there is an insignificant difference in efficacy of 4.4%.
Figure 4.
IAC amplification with low-level N. gonorrhoeae. The figure shows the internal amplification control amplified in dilutions of increasing concentrations of N. gonorrhoeae DNA. The amplification lines that do not cross the thick line are negative due to larger amounts of N. gonorrhoeae DNA.
Figure 5.
Dilution series of N. gonorrhoeae DNA using two parallels from master mixes with and without IAC. There was no significant difference in the amplification of N. gonorrhoeae DNA between the two master mixes as all four parallels are approximately equal. The mastermix without IAC shows a slightly higher ΔRn than the master mix with IAC.
One N. gonorrhoeae reference strain (ATCC 19424) was aliquoted and stored at −20°C and thawed for DNA purification and PCR. At the same time a commercially purified DNA sample (Tebu-bio) was aliquoted and stored at −20°C. These two samples were run 7 different days over a 2-week period with an average Ct value for the ATCC 19424 strain of 33.98 and a SD of 0.8. For the commercially purified DNA, the Ct was 36 on average with a SD of 0.4.
The limit of detection for the commercially isolated and quantified N. gonorrhoeae DNA was determined to 7.5 Geq/PCR reaction in 5:5 parallel reactions. In 3:5 parallels, we detected 1.5 Geq/PCR reaction. When evaluating a dilution series of N. gonorrhoeae spiked UTM-RT medium, we found the colony-forming units/ml UTM-RT to be 80 in one experiment and 53 the next experiment, which in average translates to 0.13 colony-forming units/μl DNA eluate, given a 100% recovery in the DNA isolation. This was based on one experiment where the dilutions were plated in duplicates, and the PCR was run in parallels.
Discussion
Our results show that the real-time porA pseudogene PCR, developed in the present study, was sensitive and specific enough to detect all of the 210 highly different N. gonorrhoeae international reference strains and clinical isolates examined in the present study, ie, had a sensitivity of 100%. In addition, the detection limit was low (7.5 Geq/PCR reaction).
Regarding the specificity of the method, in Figure 1 a partial alignment of N. gonorrhoeae porA consensus pseudogene (composite of 87 previously sequenced N. gonorrhoeae isolates34) and the corresponding region of the N. meningitidis porA gene show that the primers and probe are located in a highly conserved area of the porA pseudogene and that the regions are sufficiently different from the N. meningitidis porA gene to discriminate the two bacteria. Furthermore, no international reference strain or clinical isolate of any other Neisseria species was amplified. In the heterogeneous panel of other infectious agents, we included all microorganisms that we routinely analyze with PCR. In addition, we included pathogens and commensal nonpathogens available in our collection of isolates. The choice of commensals aimed to cover a representative selection of microorganisms that can be derived from all main test sites used for gonorrhea sampling; ie, cervix, urethra, oropharynx, and anorectum. None of these non-Neisseria agents were amplified by the porA pseudogene PCR. Consequently, the assay comprised 100% specificity.
Several commercial suppliers have developed NAATs for N. gonorrhoeae, but many of these as well as the developed in-house methods suffer from too-low specificity.19,36,43,44 This problem has been due to limited sequencing of equivalent genes in commensal Neisseria species and the overall great homology between most Neisseria species. Recent work on sequencing of the entire porA pseudogene in N. gonorrhoeae34 has facilitated the discovery of this gene as a promising target for highly sensitive and specific molecular diagnostics of N. gonorrhoeae.
The porA gene exists as a highly conserved, nonexpressed pseudogene in N. gonorrhoeae and is therefore not subjected to a high evolutionary selective pressure for changes as is the case for the expressed N. meningitidis porA gene.34 This is an advantage, as it might limit the risk of false negatives because of mutations in the gene in the future. On the other hand, bacteria are not believed to retain unnecessary genes, and this may predict a loss of the porA pseudogene from the N. gonorrhoeae genome in an evolutionary time perspective.45 However, the porA pseudogene seems to have been retained since N. gonorrhoeae and N. meningitidis diverged into two different species from their common ancestor.33,46 In the present study, we tested isolates cultured during a time period spanning from the 1960s to the present date, which at least demonstrates a certain degree of stability of the porA pseudogene. There might even be an unidentified reason for keeping this gene.
Previously, we have tried numerous other genes as possible targets for a N. gonorrhoeae-specific PCR, such as traG,47 traH,47 dcmG,48 carbonic anhydrase,49 and pJD1.26 However, all these PCRs have either amplified commensal Neisseria spp. genes or failed to identify some N. gonorrhoeae isolates. The results of the present study and the increasing data regarding the characteristics of the porA pseudogene,33,34 strongly support that so far the porA pseudogene is the most suitable target for N. gonorrhoeae diagnostic NAATs.
One major advantage with the real-time diagnostic assay developed in the present study is that the TaqMan chemistry can be used on most existing real-time PCR platforms, making it more easily accessible than FRET (free resonance energy transfer)-probe chemistry, which was used by Whiley et al.27,31,32,36 Furthermore, because there is a significant difference (140 bp) in the size of the N. gonorrhoeae porA pseudogene amplicon and the IAC, the different amplicons are easily distinguished by melting-point analysis using, for example, SYBR Green I fluorescence in non-probe-based real-time PCR or on agarose gel electrophoresis after conventional PCR (data not shown). A PCR with our primers used without the probe was able to discriminate against N. meningitidis and commensal Neisseria (data not shown). Consequently, the PCR developed in the present study can readily be adapted and optimized to be used also in resource-poor settings, in which probe-based real-time PCR systems are not accessible or afforded.
In conclusion, the N. gonorrhoeae porA pseudogene real-time PCR developed in the present study is highly sensitive, specific, robust, rapid, and reproducible, making it suitable for diagnosis of N. gonorrhoeae. Comparison against culturing on samples taken from several different collection sites is necessary for further evaluation. We are currently doing this comparison in collaboration with external laboratories.
Acknowledgments
We thank Helen Palmer for providing us with additional reference strains and isolates of N. gonorrhoeae and Lene Berthelsen for providing isolates of both N. gonorrhoeae and commensal Neisseria sp. We thank David Whiley for recommending the porA pseudogene as a suitable target for N. gonorrhoeae diagnostics.
Footnotes
This work was supported in full by the Department of Microbiology at the University Hospital of North Norway.
The present study was performed at the Department of Microbiology and Infection Control, University Hospital of North Norway, Tromsø, Norway. The project was also funded by the same department.
References
- Graver MA, Wade JJ. Survival of Neisseria gonorrhoeae isolates of different auxotypes in six commercial transport systems. J Clin Microbiol. 2004;42:4803–4804. doi: 10.1128/JCM.42.10.4803-4804.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bassiri M, Mårdh PA, Domeika M. Multiplex AMPLICOR PCR screening for Chlamydia trachomatis and Neisseria gonorrhoeae in women attending non-sexually transmitted disease clinics. The European Chlamydia Epidemiology Group. J Clin Microbiol. 1997;35:2556–2560. doi: 10.1128/jcm.35.10.2556-2560.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmale JD, Martin JE, Jr, Domescik G. Observations on the culture diagnosis of gonorrhea in women. JAMA. 1969;210:312–314. [PubMed] [Google Scholar]
- Knapp JS. Historical perspectives and identification of Neisseria and related species. Clin Microbiol Rev. 1988;1:415–431. doi: 10.1128/cmr.1.4.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapsall JW. Antibiotic resistance in Neisseria gonorrhoeae. Clin Infect Dis. 2005;41(Suppl 4):S263–S268. doi: 10.1086/430787. [DOI] [PubMed] [Google Scholar]
- Guibourdenche M, Popoff MY, Riou JY. Deoxyribonucleic acid relatedness among Neisseria gonorrhoeae, N. meningitidis, N. lactamica, N. cinerea and “Neisseria polysaccharea”. Ann Inst Pasteur Microbiol. 1986;137B:177–185. doi: 10.1016/s0769-2609(86)80106-5. [DOI] [PubMed] [Google Scholar]
- Perrin A, Nassif X, Tinsley C. Identification of regions of the chromosome of Neisseria meningitidis and Neisseria gonorrhoeae which are specific to the pathogenic Neisseria species. Infect Immun. 1999;67:6119–6129. doi: 10.1128/iai.67.11.6119-6129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrin A, Bonacorsi S, Carbonnelle E, Talibi D, Dessen P, Nassif X, Tinsley C. Comparative genomics identifies the genetic islands that distinguish Neisseria meningitidis, the agent of cerebrospinal meningitis, from other Neisseria species. Infect Immun. 2002;70:7063–7072. doi: 10.1128/IAI.70.12.7063-7072.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton HL, Dominguez NM, Schwartz KJ, Hackett KT, Dillard JP. Neisseria gonorrhoeae secretes chromosomal DNA via a novel type IV secretion system. Mol Microbiol. 2005;55:1704–1721. doi: 10.1111/j.1365-2958.2005.04521.x. [DOI] [PubMed] [Google Scholar]
- Davidsen T, Tonjum T. Meningococcal genome dynamics. Nat Rev Microbiol. 2006;4:11–22. doi: 10.1038/nrmicro1324. [DOI] [PubMed] [Google Scholar]
- Geraats-Peters CW, Brouwers M, Schneeberger PM, van der Zanden AG, Bruisten SM, Weers-Pothoff G, Boel CH, van den Brule AJ, Harmsen HG, Hermans MH. Specific and sensitive detection of neisseria gonorrhoeae in clinical specimens by real-time PCR. J Clin Microbiol. 2005;43:5653–5659. doi: 10.1128/JCM.43.11.5653-5659.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RE, Newhall WJ, Papp JR, Knapp JS, Black CM, Gift TL, Steece R, Markowitz LE, Devine OJ, Walsh CM, Wang S, Gunter DC, Irwin KL, DeLisle S, Berman SM. Screening tests to detect Chlamydia trachomatis and Neisseria gonorrhoeae infections–2002. MMWR Recomm Rep. 2002;51:1–38. [PubMed] [Google Scholar]
- Kellogg ND, Baillargeon J, Lukefahr JL, Lawless K, Menard SW. Comparison of nucleic acid amplification tests and culture techniques in the detection of Neisseria gonorrhoeae and Chlamydia trachomatis in victims of suspected child sexual abuse. J Pediatr Adolesc Gynecol. 2004;17:331–339. doi: 10.1016/j.jpag.2004.07.006. [DOI] [PubMed] [Google Scholar]
- Koumans EH, Black CM, Markowitz LE, Unger E, Pierce A, Sawyer MK, Papp JR. Comparison of methods for detection of Chlamydia trachomatis and Neisseria gonorrhoeae using commercially available nucleic acid amplification tests and a liquid pap smear medium. J Clin Microbiol. 2003;41:1507–1511. doi: 10.1128/JCM.41.4.1507-1511.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livengood CH, III, Wrenn JW. Evaluation of COBAS AMPLICOR (Roche): accuracy in detection of Chlamydia trachomatis and Neisseria gonorrhoeae by coamplification of endocervical specimens. J Clin Microbiol. 2001;39:2928–2932. doi: 10.1128/JCM.39.8.2928-2932.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luijt DS, Bos PA, van Zwet AA, Voorst Vader PC, Schirm J. Comparison of COBAS AMPLICOR Neisseria gonorrhoeae PCR, including confirmation with N. gonorrhoeae-specific 16S rRNA PCR, with traditional culture. J Clin Microbiol. 2005;43:1445–1447. doi: 10.1128/JCM.43.3.1445-1447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lum G, Freeman K, Nguyen NL, Limnios EA, Tabrizi SN, Carter I, Chambers IW, Whiley DM, Sloots TP, Garland SM, Tapsall JW. A cluster of culture positive gonococcal infections but with false negative cppB gene based PCR. Sex Transm Infect. 2005;81:400–402. doi: 10.1136/sti.2004.013805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahony JB, Luinstra KE, Tyndall M, Sellors JW, Krepel J, Chernesky M. Multiplex PCR for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in Genitourinary specimens. J Clin Microbiol. 1995;33:3049–3053. doi: 10.1128/jcm.33.11.3049-3053.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer HM, Mallinson H, Wood RL, Herring AJ. Evaluation of the specificities of five DNA amplification methods for the detection of Neisseria gonorrhoeae. J Clin Microbiol. 2003;41:835–837. doi: 10.1128/JCM.41.2.835-837.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapsall JW, Limnios EA, Nguyen NL, Carter I, Lum G, Freeman K, Tabrizi SN, Garland SM, Whiley DM, Sloots TP, Chambers IW. Cryptic-plasmid-free gonococci may contribute to failure of cppB gene-based assays to confirm results of BD ProbeTEC PCR for identification of Neisseria gonorrhoeae. J Clin Microbiol. 2005;43:2036–2037. doi: 10.1128/JCM.43.4.2036-2037.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Pol B, Ferrero DV, Buck-Barrington L, Hook E, III, Lenderman C, Quinn T, Gaydos CA, Lovchik J, Schachter J, Moncada J, Hall G, Tuohy MJ, Jones RB. Multicenter evaluation of the BDProbeTec ET System for detection of Chlamydia trachomatis and Neisseria gonorrhoeae in urine specimens, female endocervical swabs, and male urethral swabs. J Clin Microbiol. 2001;39:1008–1016. doi: 10.1128/JCM.39.3.1008-1016.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Doornum GJ, Schouls LM, Pijl A, Cairo I, Buimer M, Bruisten S. Comparison between the LCx Probe system and the COBAS AMPLICOR system for detection of Chlamydia trachomatis and Neisseria gonorrhoeae infections in patients attending a clinic for treatment of sexually transmitted diseases in Amsterdam, The Netherlands. J Clin Microbiol. 2001;39:829–835. doi: 10.1128/JCM.39.3.829-835.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centers for Disease Control and Prevention (CDC) Recall of LCx Neisseria gonorrhoeae assay and implications for laboratory testing for N. gonorrhoeae and Chlamydia trachomatis. MMWR Morb Mortal Wkly Rep. 2002;51:709. [PubMed] [Google Scholar]
- Smith DW, Tapsall JW, Lum G. Guidelines for the use and interpretation of nucleic acid detection tests for Neisseria gonorrhoeae in Australia: a position paper on behalf of the Public Health Laboratory Network. Commun Dis Intel. 2005;29:358–365. [PubMed] [Google Scholar]
- Farrell DJ. Evaluation of AMPLICOR Neisseria gonorrhoeae PCR using cppB nested PCR and 16S rRNA PCR. J Clin Microbiol. 1999;37:386–390. doi: 10.1128/jcm.37.2.386-390.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korch C, Hagblom P, Öhman H, Göransson M, Normark S. Cryptic plasmid of Neisseria gonorrhoeae: complete nucleotide sequence and genetic organization. J Bacteriol. 1985;163:430–438. doi: 10.1128/jb.163.2.430-438.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley DM, LeCornec GM, Mackay IM, Siebert DJ, Sloots TP. A real-time PCR assay for the detection of Neisseria gonorrhoeae by LightCycler. Diagn Microbiol Infect Dis. 2002;42:85–89. doi: 10.1016/s0732-8893(01)00326-1. [DOI] [PubMed] [Google Scholar]
- Chaudhry U, Saluja D. Detection of Neisseria gonorrhoeae by PCR using orf1 gene as target. Sex Transm Infect. 2002;78:72. doi: 10.1136/sti.78.1.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boel CH, van Herk CM, Berretty PJ, Onland GH, van den Brule AJ. Evaluation of conventional and real-time PCR assays using two targets for confirmation of results of the COBAS AMPLICOR Chlamydia trachomatis/Neisseria gonorrhoeae test for detection of Neisseria gonorrhoeae in clinical samples. J Clin Microbiol. 2005;43:2231–2235. doi: 10.1128/JCM.43.5.2231-2235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana C, Favaro M, Cicchetti O, Minelli S, Pistoia ES, Favalli C. Performance of strand displacement amplification assay in the detection of Chlamydia trachomatis and Neisseria gonorrhoeae. Jpn J Infect Dis. 2005;58:283–288. [PubMed] [Google Scholar]
- Whiley DM, Sloots TP. Comparison of three in-house multiplex PCR assays for the detection of Neisseria gonorrhoeae and Chlamydia trachomatis using real-time and conventional detection methodologies. Pathology. 2005;37:364–370. doi: 10.1080/00313020500254552. [DOI] [PubMed] [Google Scholar]
- Whiley DM, Buda PP, Freeman K, Pattle NI, Bates J, Sloots TP. A real-time PCR assay for the detection of Neisseria gonorrhoeae in genital and extragenital specimens. Diagn Microbiol Infect Dis. 2005;52:1–5. doi: 10.1016/j.diagmicrobio.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Feavers IM, Maiden MC. A gonococcal porA pseudogene: implications for understanding the evolution and pathogenicity of Neisseria gonorrhoeae. Mol Microbiol. 1998;30:647–656. doi: 10.1046/j.1365-2958.1998.01101.x. [DOI] [PubMed] [Google Scholar]
- Unemo M, Norlén O, Fredlund H. The porA pseudogene of Neisseria gonorrhoeae–low level of genetic polymorphism and a few, mainly identical, inactivating mutations. APMIS. 2005;113:410–419. doi: 10.1111/j.1600-0463.2005.apm_206.x. [DOI] [PubMed] [Google Scholar]
- Derrick JP, Urwin R, Suker J, Feavers IM, Maiden MC. Structural and evolutionary inference from molecular variation in Neisseria porins. Infect Immun. 1999;67:2406–2413. doi: 10.1128/iai.67.5.2406-2413.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiley DM, Buda PJ, Bayliss J, Cover L, Bates J, Sloots TP. A new confirmatory Neisseria gonorrhoeae real-time PCR assay targeting the porA pseudogene. Eur J Clin Microbiol Infect Dis. 2004;23:705–710. doi: 10.1007/s10096-004-1170-0. [DOI] [PubMed] [Google Scholar]
- Van Dyck E, Meheus AZ, Piot P. Geneva: World Health Organization (WHO); Laboratory diagnosis of sexually transmitted diseases. 1999:135 pp. [Google Scholar]
- Sneed J. Processing and interpretation of upper respiratory tract specimens. Isenberg DH, editor. Washington, DC: American Society for Microbiology; 1992:14.7–14.21. [Google Scholar]
- Christensen JJ, Ursing J, Bruun B. Genotypic and phenotypic relatedness of 80 strains of Branhamella catarrhalis of worldwide origin. FEMS Microbiol Lett. 1994;119:155–159. doi: 10.1111/j.1574-6968.1994.tb06882.x. [DOI] [PubMed] [Google Scholar]
- Martin JE, Jr, Lester A. Transgrow, a medium for transport and growth of Neisseria gonorrhoeae and Neisseria meningitidis. HSMHA Health Rep. 1971;86:30–33. [PMC free article] [PubMed] [Google Scholar]
- Rutledge RG, Cote C. Mathematics of quantitative kinetic PCR and the application of standard curves. Nucleic Acids Res. 2003;31:e93. doi: 10.1093/nar/gng093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J, Jeffries AC, Nelson KE, Eisen JA, Ketchum KA, Hood DW, Peden JF, Dodson RJ, Nelson WC, Gwinn ML, DeBoy R, Peterson JD, Hickey EK, Haft DH, Salzberg SL, White O, Fleischmann RD, Dougherty BA, Mason T, Ciecko A, Parksey DS, Blair E, Cittone H, Clark EB, Cotton MD, Utterback TR, Khouri H, Qin H, Vamathevan J, Gill J, Scarlato V, Masignani V, Pizza M, Grandi G, Sun L, Smith HO, Fraser CM, Moxon ER, Rappuoli R, Venter JC. Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science. 2000;287:1809–1815. doi: 10.1126/science.287.5459.1809. [DOI] [PubMed] [Google Scholar]
- Bachmann LH, Desmond RA, Stephens J, Hughes A, Hook EW., III Duration of persistence of gonococcal DNA detected by ligase chain reaction in men and women following recommended therapy for uncomplicated gonorrhea. J Clin Microbiol. 2002;40:3596–3601. doi: 10.1128/JCM.40.10.3596-3601.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diemert DJ, Libman MD, Lebel P. Confirmation by 16S rRNA PCR of the COBAS AMPLICOR CT/NG test for diagnosis of Neisseria gonorrhoeae infection in a low-prevalence population. J Clin Microbiol. 2002;40:4056–4059. doi: 10.1128/JCM.40.11.4056-4059.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- Vazquez JA, de la Fuente L, Berron S, O’Rourke M, Smith NH, Zhou J, Spratt BG. Ecological separation and genetic isolation of Neisseria gonorrhoeae and Neisseria meningitidis. Curr Biol. 1993;3:567–572. doi: 10.1016/0960-9822(93)90001-5. [DOI] [PubMed] [Google Scholar]
- Dillard JP, Seifert HS. A variable genetic island specific for Neisseria gonorrhoeae is involved in providing DNA for natural transformation and is found more often in disseminated infection isolates. Mol Microbiol. 2001;41:263–277. doi: 10.1046/j.1365-2958.2001.02520.x. [DOI] [PubMed] [Google Scholar]
- Stein DC, Gunn JS, Radlinska M, Piekarowicz A. Restriction and modification systems of Neisseria gonorrhoeae. Gene. 1995;157:19–22. doi: 10.1016/0378-1119(94)00649-d. [DOI] [PubMed] [Google Scholar]
- Chirica LC, Elleby B, Jonsson BH, Lindskog S. The complete sequence, expression in Escherichia coli, purification and some properties of carbonic anhydrase from Neisseria gonorrhoeae. Eur J Biochem. 1997;244:755–760. doi: 10.1111/j.1432-1033.1997.00755.x. [DOI] [PubMed] [Google Scholar]