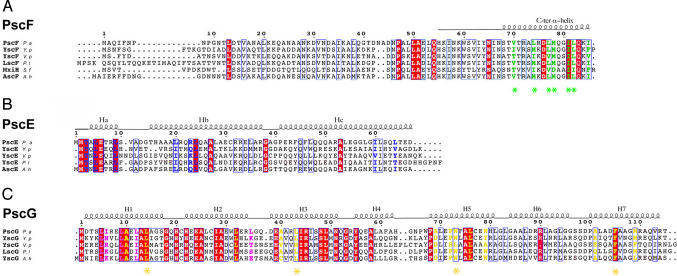
Fig. 1.
Structure-based sequence alignment of T3SS needle proteins and their cytoplasmic partners from diverse pathogens. (A) PscF from P. aeruginosa (causative agent of nosocomial infections), YscF from Y. pestis (bubonic plague), YscF from Vibrio parahaemolyticus (gastrointestinal illness), LscF from Photorhabdus luminescens (insect pathogen), MxiH from S. flexneri (bacillary dysentery), and AscF from Aeromonas hydrophila (respiratory illness in humans, reptiles, and birds). Residues mutated in this study and shown to be involved in polymerization and needle stability are highlighted with stars, whereas those which interact with PscG are shown in green. (B) PscE homologs from bacteria described above share a major homology region within their N-terminal domains. PscE residues which interact with PscG are shown in blue. (C) PscG homologs share high conservation within residues which interact with PscE (purple) and PscF (yellow). Residues mutated to Ser in this study are indicated with stars.