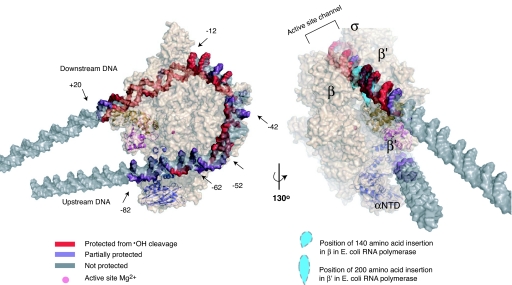
Fig. 3.
Model of the intermediate complex I1 preceding formation of the transcriptionally competent RPo. The conformation of RNAP (beige) is based on the x-ray crystal structure of the homologous T. thermophilus enzyme [Protein Data Bank ID code 1IW7 (16); T. thermophilus β′ residues 138–452, absent in E. coli (28), are not shown]. The modeled position of promoter DNA in I1 is based on DNase I (6), ·OH and KMnO4 footprinting and relevant crystal structures. The two views of I1 demonstrate the agreement between the model and the experimentally determined strong and moderate protection (red and purple, respectively) of the DNA backbone (gray) from ·OH cleavage (see Fig. 1). Strong protection seen at −12 to −19 likely involves the nonconserved region of E. coli σ70 not present in σA of T. thermophilus. Domains in E. coli subunits β and β′ (represented as blue teardrops, missing in T. thermophilus) are positioned at the sites of their insertion in the T. thermophilus sequence; these likely protect DNA downstream of +10 from ·OH cleavage. The upstream surface groove formed by β′ and the NTD of α, and the mobile downstream jaw (E. coli β′ residues 913-1262) and upstream DNA clamp (E. coli β′ residues 808–912), are highlighted in cartoon representations in dark blue, magenta, and gold, respectively. This figure was created using PyMol (41).