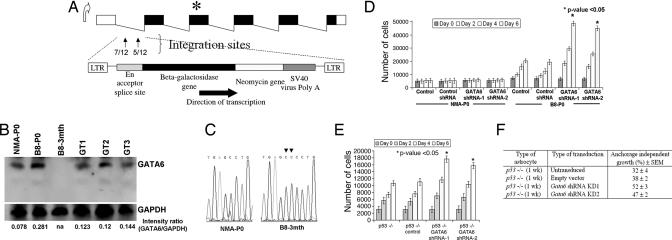
Fig. 2.
Characteristics of gene-trapped astrocytes. (A) Schematic of the murine Gata6 transcript isoform expressed in the CNS. Integrations of the retroviral gene-trap vector were identified within the first intron of Gata6 and in the same orientation as the endogenous promoter. The number of integrations per site is shown in the first intron. ∗ represents the map position of a homozygous frameshift mutation identified in exon 3 of B8–3mth astrocytes. (B) Western blot analyses on protein lysates isolated from primary astrocyte cultures, with 40 μg of total protein lysate loaded per lane. Absent Gata6 expression is noted in B8–3mth astrocytoma cells (consistent with findings in Fig. 1 B and C). GAPDH was used as a positive control for loading and normalization of the densitometric analyses with Fluor Chem software. Gata6 expression was reduced between ≈50% and 58% among the three B8-P0 gene-trapped clones analyzed in detail (GT1, GT2, and GT3). (C) Chromatograms demonstrating a 1641_1642InsCC mutation in exon 3 of Gata6 in the B8–3mth astrocytes encoding the DNA binding domain. (D) Gata6 expression was constitutively knocked-down in NMA and B8-P0 astrocytes by ≈90% compared with controls by Western blot analysis (data not shown), with two shRNAs (mapping to murine exons 2 and 4) expressed from a stably integrated pSIREN-RetroQ vector. Parental or NMA and B8-P0 astrocytes engineered with a negative shRNA vector (control) were used as negative controls for the MTT proliferation assays. Gata6 knockdown induced a significant (∗, P < 0.05) proliferation advantage after day 2, only in the B8-P0 astrocytes and not in NMA. (E) Gata6 knockdown in homozygous null murine astrocytes induced a proliferative advantage (P < 0.05) and increased anchorage-independent growth in soft agar (F) compared with parental p53−/− astrocytes or those transduced with a negative shRNA vector (control).