Table 1.
The A. nidulans CSN subunits
| CSN subunit | A. nidulans protein no. | A. nidulans protein name | Domain | e value | % amino acid identities to: |
Positions of introns and PCI/MPN domain | Coding regions, bp | Protein, kDa | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| hs | at | sp | sc | ||||||||
| CSN1 | AN1491 | CsnA | PCI | 2.1 e−07 | 33.3 | 38.6 | 27.0 | — | 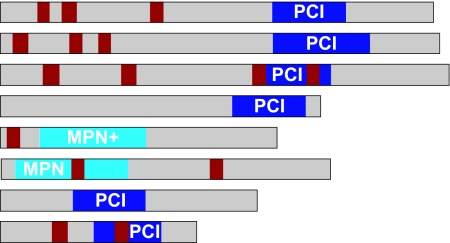 |
1,497 | 55.7 |
| CSN2 | AN4783 | CsnB | PCI | 1.5 e−16 | 51.1 | 48.1 | 46.1 | — | 1,521 | 58.1 | |
| CSN3 | AN5798 | CsnC | PCI | 6.0 e+01 | 26.0 | 23.4 | 20.1 | — | 1,491 | 55.2 | |
| CSN4 | AN1539 | CsnD | PCI | 1.1 e−03 | 38.5 | 41.2 | 19.4 | — | 1,227 | 44.9 | |
| CSN5 | AN2129 | CsnE | MPN+ | 6.6 e−37 | 55.2 | 54.4* | 38.8 | 27.8 | 1,008 | 37.8 | |
| CSN6 | AN2233 | CsnF | MPN | 3.9 e−01 | 31.1 | 29.9* | — | — | 1,161 | 42.1 | |
| CSN7 | AN3623 | CsnG | PCI | 7.6 e−15 | 28.8* | 29.5 | 27.0 | — | 984 | 35.3 | |
| CSN8 | AN10208 | CsnH | PCI | 1.4 e+02 | 22.4 | 18.9 | — | — | 639 | 23.9 | |
CsnA–CsnH contain PCI and MPN domains (SMART-ID: SM00088/SM00232); poor E values are indicated by italics. Percentages of amino acid identities to the sequences of Homo sapiens (hs), Arabidopsis thaliana (at), Schizosaccharomyces pombe (sp), and S. cerevisiae (sc) (26) are given (sequence IDs are in SI Table 3). Positions of PCI/MPN (blue) and introns (red) within coding regions are indicated.
*Isoforms: hsCSN7B (27.8%), atCSN5B (53.2%), and atCSN6B (31.1%).
