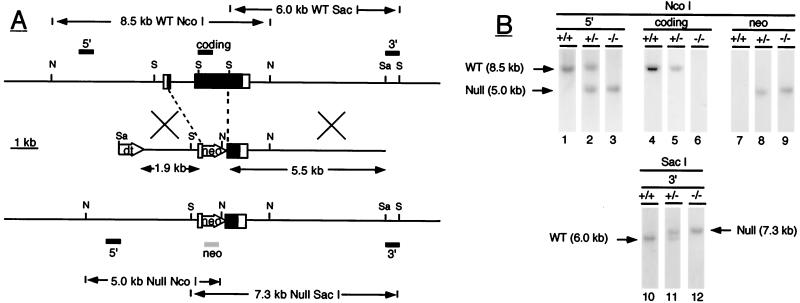
Figure 1.
(A) Inactivation of the ALS gene by homologous recombination. The wild-type ALS gene is shown at the top with exons represented by boxes (coding regions are closed; noncoding regions are open) and with intron and flanking regions represented by horizontal lines. Locations of the restriction endonuclease sites for NcoI (N), SacI (S), and SalI (Sa) are shown by horizontal bars, and positions of the various ALS probes used (5′, coding and 3′) by closed rectangles above the gene. In wild-type DNA, the 5′ and coding ALS probes detect an 8.5-kb NcoI fragment whereas the 3′ probe detects a 6.0-kb SacI fragment. The middle schematic shows the targeting vector containing the ALS genomic fragments and the neomycin phosphotransferase (neo) and diphtheria toxin (dt) genes. Regions of homology between the vector and the wild-type gene are indicated by crossed bars. The null allele produced by homologous recombination is represented at the bottom. In null ALS DNA, the 5′ ALS and neo probes (shown as a shaded rectangle under the null allele) detect a 5.0-kb NcoI fragment whereas the 3′ ALS probe detects a 7.3-kb SacI fragment. (B) Identification of null ALS mice by Southern blotting. Genomic DNA was digested with the restriction enzyme NcoI (lanes 1–9) or SacI (lanes 10–12) and was analyzed by Southern blotting with the indicated DNA probes. The hybridization pattern obtained with each probe is shown for wild-type (+/+), heterozygous (+/−), and null (−/−) ALS mice. DNA fragments corresponding to the wild-type (wt) and null allele (null) are indicated by arrows.