Fig. 2. Conservation of coding information for CtBPL-specific C-terminus.
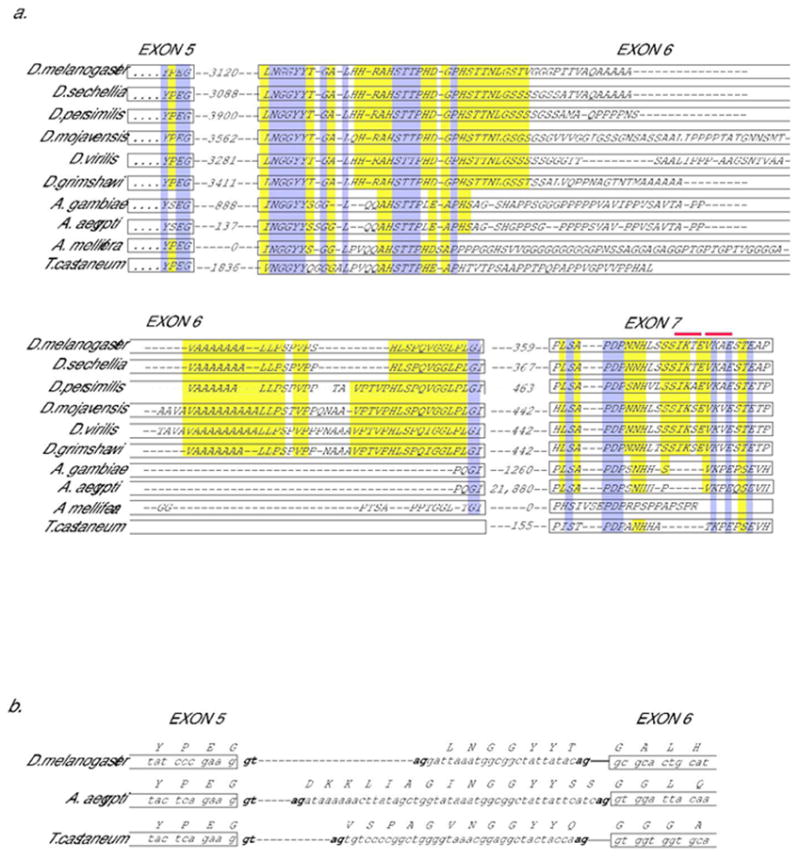
A. Peptide coding information present in dipterans, bee, and beetle genomic sequences homologous to alternatively spliced exon 6 and 7 in Drosophila melanogaster encoding CtBP “tail” region. Conceptual translations of genomic sequences are shown below sequence of CtBPL, in which YPEG represents the end of the exon 5 coding sequence for the CtBPL isoform. Predicted intron size in nucleotides is indicated between exons. The introns in Apis mellifera have apparently been eliminated. Dark gray (purple) shading indicates widely conserved sequences; light gray (yellow) shading partially conserved sequences. Possible sumoylation sites (I/V K X E) are indicated by gray (red) bars above exon 7 residues. An alternative splice acceptor site 5’ of the junction shown for exon 7 would produce an mRNA encoding an additional VSSQS motif at the beginning of exon 7; this sequence is not conserved outside of Drosophila, unlike the case shown in 2B. B. The cDNA sequences reported for D. melanogaster CtBPL contain alternative splice acceptor sites for the 5’ end of exon 6; a sequence isolated from adult head uses a downstream acceptor site (NP_001014617), while a different sequence isolated from embryo uses a more upstream acceptor ((Sutrias-Grau and Arnosti, 2004)) incorporating the residues LNGGYYT. This portion of the protein is evolutionarily conserved and contains appropriate splice acceptor sequences both 5’ and 3’ of region, thus alternative splicing may be a conserved feature here as well.
