Abstract
Control of cellular dimensions and cell symmetry are critical for development and differentiation. Here we provide evidence that the putative Rho-GAP Rga4p of Schizosaccharomyces pombe controls cellular dimensions. rga4Δ cells are wider in diameter and shorter in length, whereas Rga4p overexpression leads to reduced diameter of the growing cell tip. Consistent with a negative role in cell growth control, Rga4p protein localizes to the cell sides in a “corset” pattern, and to the nongrowing cell tips. Additionally, rga4Δ cells show an altered growth pattern similar to that observed in mutants of the formin homology protein For3p. Consistent with these observations, Rga4p is required for normal localization of For3p and for normal distribution of the actin cytoskeleton. We show that different domains of the Rga4p protein mediate diverse morphological functions. The C-terminal GAP domain mediates For3p localization to the cell tips and maintains cell diameter. Conversely, overexpression of the N-terminal LIM homology domain of Rga4p promotes actin cable formation in a For3p-dependent manner. Our studies indicate that Rga4p functionally interacts with For3p and has a novel function in the control of cell diameter and cell growth.
INTRODUCTION
Control of cell morphology is critical for cell differentiation and for tissue and organ development. An alteration of cell shape and cell size is observed during neurogenesis and cardiac development in animal cells (Nelson, 2003; Sano and Schneider, 2003) and in the genesis of root hairs, pollen tubes, and vascular elements in plant cells (Smith, 2003). The spatial control of cytoskeletal structures is also essential for the asymmetric inheritance of regulatory proteins and asymmetric cell division, which ensures the ordered development of multicellular organisms (Bardin et al., 2004).
The fission yeast Schizosaccharomyces pombe has a well-defined cylindrical shape and grows in a polarized manner from the cell ends. Polarized cell growth is strictly cell cycle regulated. After cytokinesis, the two daughter cells grow in a monopolar manner by initiating growth at the old tips in a process termed Old End Take Off. Once a minimal cell length has been reached, the cells activate bipolar growth by initiating growth from the newly formed tips (New End Take Off [NETO]; Streiblova and Wolf, 1972; Nurse, 1975; Mitchison and Nurse, 1985). Thus, the pattern of cell growth is generally symmetrical in the two daughter cells.
The regular shape of the S. pombe cell has facilitated the study of morphology control mechanisms. Several genetic screens have led to the identification of numerous genes involved in the control of cell polarity, and the actin and microtubule cytoskeleton (Verde, 1998; Chang and Verde, 2004). Mutants in these genes become round, displaying loss of polarity, or become bent and branched, if the molecular mechanisms that mark the site of cell growth are defective.
Conversely, there has been very little study on the control cell diameter. The overall diameter of the S. pombe cell is on an average ∼3.5 μm (Mitchinson, 1957). Cell diameter is maintained constant during the cell cycle with little variation between individuals (Streiblova and Wolf, 1972). Studies have shown that cell diameter increases in older cells, during exposure to low osmolarity, and in diploid cells (Mitchinson, 1970; Kubitschek and Clay, 1986). Studies in Arabidopsis thaliana have shown that the root hair growth and diameter at the tip is controlled by RHD4 (Galway et al., 1999) and is dependent on proper actin organization (Preuss et al., 2004). The well-defined shape of fission yeast in combination with its maintenance of diameter throughout the cell cycle makes it an ideal system for studying cell diameter control.
Cell morphogenesis in fission yeast is regulated by signaling pathways that are conserved in higher eukaryotic cells (Chang and Peter, 2003; Chang and Verde, 2004). These pathways include the conserved Cdc42p Rho GTPase (Miller and Johnson, 1994), its regulators and its effectors, and five other Rho GTPases Rho1p–Rho5p (Nakano and Mabuchi, 1995; Arellano et al., 1996; Nakano et al., 1997). These GTPases are regulated by Rho-GAPs that promote GTP hydrolysis. There are eight putative Rho-GAPs in S. pombe, Rga1p–Rga8p (Nakano et al., 2001). Rga1p has been shown to have a function in the control of cell morphology (Nakano et al., 2001), whereas Rga5p has a role in cell integrity (Calonge et al., 2003). Studies also suggest that Rga8p has a negative role in cell growth control (Yang et al., 2003). The exact function of the other Rho-GAPs is still unknown.
Formins are a group of Rho-GTPase activated, conserved proteins that are required for basic cellular processes such as cell adhesion, cell motility, cell polarity, and cell division (as reviewed in Zigmond, 2004; Higgs, 2005; and Faix and Grosse, 2006; Kovar, 2006) and are essential for efficient incorporation of actin-profilin at the barbed end of actin filaments (Kovar et al., 2003 and 2006; Romero et al., 2004). In fission yeast, the formin For3p regulates actin cable formation and for3Δ mutants show a defective growth pattern, with loss of symmetric division and polarized growth, and reduction in the number of actin cables (Feierbach and Chang, 2001). Recent studies have shown that For3p binds at the cortical tips transiently and promotes formation of short actin filaments that are then bundled into cables (Martin and Chang, 2006). The mechanism by which For3p binds to the cell tip is still not well understood. For3p depends on the linker protein Tea4p/Wsh3p for normal localization to both cell ends (Martin et al., 2005; Tatebe et al., 2005). This interaction with Tea4p/Wsh3p promotes the association of For3p with Tea1p, a kelch repeat containing protein that functions as a marker at the cell tips (Snell and Nurse, 1994; Mata and Nurse, 1997; Behrens and Nurse, 2002; Kim et al., 2003). For3p is localized to only one tip in tea4Δ/wsh3Δ mutants (Martin et al., 2005).
In this report we characterize the function of fission yeast Rga4p in the control of cell morphogenesis. Our observations indicate that Rga4p is a multidomain protein with several functions and is required for the control of cell diameter, for maintenance of cell growth symmetry in daughter cells, and for normal For3p localization.
MATERIALS AND METHODS
Strains and Cell Culture
S. pombe strains used in this study are listed in Table 1. All strains are isogenic to the original strain h- 972. The strains PN567 was obtained from the Paul Nurse lab, BFY81 and BFY161 from the Fred Chang lab, and CA2857 from Kazuhiro Shiozaki lab as kind gifts. Cells were cultured in minimal medium plus required supplements or yeast extract (YE) medium (http://www.hsph.harvard.edu/wolflab/Protocols/Protocols/Fission%20Yeast/Nurse%20Lab%20Manual.htm). Standard techniques were used for genetic manipulation and analysis (Moreno et al., 1991). Cells carrying the plasmid pRep3X (which contains the thiamine-repressible nmt1 promoter; Maundrell, 1993) and derivatives pRep3x-HA-rga4, pRep3x-HA-rga4ΔN, pRep3x-HA-rga4ΔGAP, and pRep3x-HA-rga4ΔLIM were grown under the following growth conditions: in minimal medium in the presence of thiamine (at 25 or 32°C), in the absence of thiamine at 32°C for 16 h, and in the absence of thiamine at 25°C for 20 h. In the presence of thiamine, protein levels of HA-Rga4p and derivatives were 1.5–2 fold greater than the levels of Rga4p-HA, expressed from the endogenous promoter at the rga4 locus. After culture in the absence of thiamine at 32°C for 16 h, protein levels of HA-Rga4p and derivatives were 20–25-fold greater than the levels of endogenous Rga4p-HA. After culture in the absence of thiamine at 25°C for 20 h, protein levels of HA-Rga4p and derivatives were 5–8-fold greater than the expression of endogenous Rga4p-HA (Das, unpublished results).
Table 1.
List of strains
| Strain | Genotype | Origin |
|---|---|---|
| PN567 | ade6-704 leu1-32 ura4-D18 h− | P. Nursea |
| FV513 | Δrga4::ura4+ ade6-704 leu1-32 ura4-D18 h+ | This study |
| BFY81 | for3-YFP::kanMX6 ura- ade- leu- h+ | F. Changb |
| FV516 | Δrga4::ura4+ ade6-704 leu1-32 ura4-D18 h+pRep3X | This study |
| FV527 | Δrga4::ura4+ orb6-HA::sup3-5 ura4-D18 ade6-704 leu1-32 | This study |
| FV531 | Δrga4::ura4+ ade6-704 leu1-32 ura4-D18pRep3X HA-rga4 h+ | This study |
| FV532 | rga4-HA:sup3-5 ade6-704 leu1-32 ura4-D18 h− | This study |
| FV542 | orb6-HA::sup3-5 ura4-D18 ade6-704 leu1-32 h+ | Wiley et al. (2003) |
| FV572 | for3-YFP::kanMX6Δrga4::ura4+ ade6- leu1- ura4-D18 | This study |
| FV573 | Δrga4::ura4+ ade6-704 leu1-32 ura4-D18 h+pRep3X HA-rga4-ΔLIM | This study |
| FV574 | Δrga4::ura4+ ade6-704 leu1-32 ura4-D18 h+pRep3X HA-rga4-ΔN | This study |
| FV575 | Δrga4::ura4+ ade6-704 leu1-32 ura4-D18 h+pRep3X HA-rga4-ΔGA | This study |
| FV584 | for3-YFP::kanMX6Δrga4::ura4+ ade6- leu1- ura4-D18pRep3X HA-rga4-ΔN | This study |
| FV585 | for3-YFP::kanMX6Δrga4::ura4+ ade6- leu1- ura4-D18pRep3X HA-rga4-ΔGAP | This study |
| FV603 | Δfor3::kanMX6 ade6-704 leu1-32 ura4-D18 h+pRep3X HA-rga4-ΔN | This study |
| FV604 | Δfor3::kanMX6 ade6-704 leu1-32 ura4-D18 h+pRep3X HA-rga4-ΔGAP | This study |
| FV605 | Δfor3::kanMX6 ade6-704 leu1-32 ura4-D18 h+pRep3X | This study |
| FV606 | Δfor3::kanMX6 ade6-704 leu1-32 ura4-D18 h+pRep3X HA-rga4 | This study |
| FV607 | pak1/shk1-HA::sup3-5 ade6-704 h− | This study |
| FV610 | pak1/shk1-HA::sup3-5Δrga4::ura4 ade- ura4-D18 | This study |
| FV743 | for3-YFP::kanMX6Δrga4::ura4+ ade6- leu1- ura4-D18pREP3X HA-rga4 | This study |
| FV759 | tea4Δ:ura4+ade6-704 leu1-32 ura4-D18pRep3X | This study |
| FV760 | tea4Δ:ura4+ade6-704 leu1-32 ura4-D18pRep3X HA-rga4 | This study |
| FV761 | tea4Δ:ura4+ade6-704 leu1-32 ura4-D18pRep3X HA-rga4-ΔGA | This study |
| FV762 | tea4Δ:ura4+ade6-704 leu1-32 ura4-D18pRep3X HA-rga4-Δ | This study |
| FV784 | rga4-GFP:kanMX ade6-704 ura4-D18 leu1-32 h+ | This study |
| FV785 | rga4-GFP:kanMX tea4Δ:ura4+ade6-704 leu1-32 ura4-D18 | This study |
| CA2301 | wsh3-GFP:ura4+ leu1-32 ura4-D18 h− | K. Shiozakic |
| FV796 | wsh3-GFP:ura4+Δrga4::ura4+ leu1-32 ura4-D18 h− | This study |
| DM3415 | myo52-GFP:kanMX ade6–210 leu1-32 ura4-D18 h− | D. McCollumd |
| FV797 | myo52-GFP:kanMX Δrga4::ura4+ ade6–210 leu1-32 ura4-D18 h− | This study |
| FV800 | ade6-704 ura4-D18 leu1-32 pRep3Xh− | This study |
| FV801 | ade6-704 ura4-D18 leu1-32pRep3X-HA-rga4 h− | This study |
| FV802 | ade6-704 ura4-D18 leu1-32pRep3X-HA-rga4-ΔN h− | This study |
| FV803 | ade6-704 ura4-D18 leu1-32pRep3X-HA-rga4-ΔGAP h− | This study |
a Yeast Genetics and Cell Biology, The Rockefeller University, New York.
b Columbia Medical Center, Department of Microbiology, Columbia University, New York.
c Section of Microbiology, Division of Biological Sciences, University of California, Davis.
d Molecular Genetics and Microbiology, University of Massachusetts Medical School, Worcester, MA.
Identification and Deletion of rga4+
The rga4 gene was identified in a two-hybrid screen for proteins that interact with the Orb6p kinase. The screen was performed as previously described (Wiley et al., 2003). The rga4 gene was cloned into the SalI and XmaI restriction sites of the pRep3X plasmid, containing the sequence for a N-terminal hemagglutinin tag, by PCR amplification from genomic DNA using the following primers: forward primer-5′-CGCGGATCCAATGAATTCGGGTACGACACTTC-3′, and reverse primer-5′-GTCCCCCCGGGTTAGGCAAAGACTTCATGTACATG-3′. To express Rga4p-HA at the endogenous level, the rga4 gene was cloned into a bluescript plasmid containing a C-terminal hemagglutinin tag and a sup3-5 marker for integration: forward primer-5′-CTGCTCGAGAAAATGAATTCG-GGTACGACACTTC-3′, and reverse primer-5′-CCCAAGCTTGGCAAAGACTTCATGTACATGATC-3′. The resulting strain (FV532) contains one copy of HA-tagged rga4 and one wild-type copy without a promoter. In addition rga4 was cloned into pFA6-GFP: forward primer: 5′-AATTGTCGACCCATCCTCCGTATTATCTGG-3′ and reverse primer: 5′-TAACCCGGGGATCCGGGCAAAGACTTCATGTACATG-3′. The PCR amplified rga4 (bases 883-3141) was cloned into pFA6-GFP digested with SalI-SmaI to construct a C-terminal GFP fusion. No linkers between rga4 gene and the GFP sequence were introduced. The construct thus generated was then digested with DraIII and transformed into PN567. G418-resistant colonies were then selected. The resulting strain contains one functional copy of rga4 fused to GFP and a truncated rga4 sequence (bases 883-3144) without an upstream promoter. To determine the functionality of the Rga4p-GFP fusion protein in the resulting strain (FV784), we examined the diameter of the strain (3.7 ± 0.2 μM) and the organization of the actin organization. The cell diameter and the actin organization of these cells were comparable to that of wild-type cells.
For construction of the Δrga4 strain (FV513), the 5′ and 3′ flanking sequences of rga4 were cloned into a bluescript plasmid flanking the ura4+ gene. This fragment was then transformed into h−ade6-704 leu1-32 ura4-D18 and Ura4+ transformants were selected. The deletion of rga4 was confirmed by Southern blot analysis. The pRep3X HA-rga4-ΔN, pRep3X HArga4-ΔGAP, and pRep3X HA-rga4-ΔLIM plasmids were made using the QuikChange II XL site-directed mutagenesis kit (Stratagene, La Jolla, CA) to make the deletions in rga4. The following primers were used for the 2250-base pair N-term. (LIM domains + Coiled Coil) deletion: forward primer: 5′-CGGGTACGACACTTCATCTGGACAATAAAGACAAGGAGAGTGGAGGG-3′, reverse primer: 5′-CCCTCCACTCTCCTTGTCTTTATTGTCCAGATGAAGTGTCGTACCCG-3′. The 716-base pair GAP deletion was made using the following primers: forward primer: 5′-CCTCACATGTCGAGTACTCCCATAGATCATGTACATGAAGTCTTTGCC-3′, and the reverse primer: 5′-GGCAAAGACTTCATGTACATGATCTATGGGAGTACTCGACATGTGAGG-3′. The 1254-base pair LIM deletion was made using the following primers: forward primer: 5′-CGGGTACGACACTTCATCTGGACGAAAGCGTTGTTAATCCTCCTCC-3′ and the reverse primer: 5′-GGAGGAGGATTAACAACGCTTTCGTCCAGATGAAGTGTCGTACCCG-3′. The QuikChange II XL protocol was followed for PCR, except the amplifications were performed with each primer in a separate reaction using half the reaction mixture. The corresponding amplifications were then combined for a final extension, and the protocol outlined in the Stratagene kit was then followed.
Immunofluorescence and Fluorescence Microscopy
Cells for immunofluorescence were grown exponentially in liquid culture for at least eight generations at densities below 107 cells/ml before the start of the experiment. Standard immunofluorescence techniques were followed (Moreno et al., 1991) after methanol fixation. The primary antibodies used for immunofluorescence were mouse monoclonal anti-HA antibody (Covance, Madison, WI), Alexa Fluor 488–conjugated anti-HA (Invitrogen, Carlsbad, CA), Tat1 anti-beta tubulin mouse mAb (a kind gift from Keith Gull, Wellcome Trust Sanger Institute, Hinxton, Cambridgeshire, United Kingdom) and a rabbit polyclonal antibody against Tea1p (kind gift from Juan Mata, Department of Biochemistry, Cambridge University, United Kingdom; Mata and Nurse, 1997). The secondary antibody was Cy3-conjugated anti-mouse antibody and Cy3-conjugated anti-rabbit antibody (Sigma, St. Louis, MO). Cells were mounted on coverslips using phosphate-buffered saline containing Slowfade Antifade (Molecular Probes, Eugene, OR) and photographed using a Zeiss Axiophot microscope (Thornwood, NY) with Openlab 3.0.2 software (Improvision, Lexington, MA) or using an Olympus fluorescence BX61 microscope (Melville, NY) equipped with Nomarski differential interference contrast (DIC) optics, a 100× objective (NA 1.35), a Roper Cool-SNAP HQ camera (Tucson, AZ), Sutter Lambda 10 + 2 automated excitation and emission filter wheels (Novato, CA), and a 175 W Xenon remote source lamp with liquid light guide. Images were acquired and processed using the Intelligent Imaging Innovations (Denver, CO) SlideBook image analysis software and prepared with Adobe Photoshop 7 (San Jose, CA). Actin staining using Alexa Fluor 488 phalloidin (Molecular Probes) was performed as described (Chang et al., 1996; Pelham and Chang, 2001). Effect of latrunculin A was analyzed on Rga4-GFP localization. Latrunculin A, 200 μM, was added to cells and pictures were taken for up to 1 h at 5-min intervals.
Analysis of Growth Pattern
Cells were grown for at least eight generations at densities below 107 cells/ml before the start of the experiment. Slides were prepared by applying 50 μl of YE medium plus 1% agar to form a thin layer on the slide surface. After allowing the medium to solidify, 1 μl of concentrated cells was placed on the medium and then coverslipped. Cells were then imaged at room temperature (∼25°C) for two generations, acquiring an image every 10 min (Rga4-GFP was analyzed in a similar manner). The images were then analyzed by measuring tip growth, as distance from the birth scar, using Openlab 3.0.2 software.
RESULTS
Rga4 Has a Role in the Control of Cellular Dimensions
In a two-hybrid screen for proteins that physically interact with the Orb6p protein kinase (Wiley et al., 2003), we identified the Rho-GAP domain-containing protein Rga4p (Nakano et al., 2001). Based on homology, rga4 (SPBC28E12.03) encodes a GTPase-activator protein for Rho-like GTPases and is similar to Rga1p and Rga2p from Saccharomyces cerevisiae (Stevenson et al., 1995; Chen et al., 1996; Nakano et al., 2001; Smith et al., 2002). To determine the cellular function of Rga4p, we deleted the rga4 gene and, as previously reported (Nakano et al., 2001), we found that rga4 is nonessential under normal growth conditions.
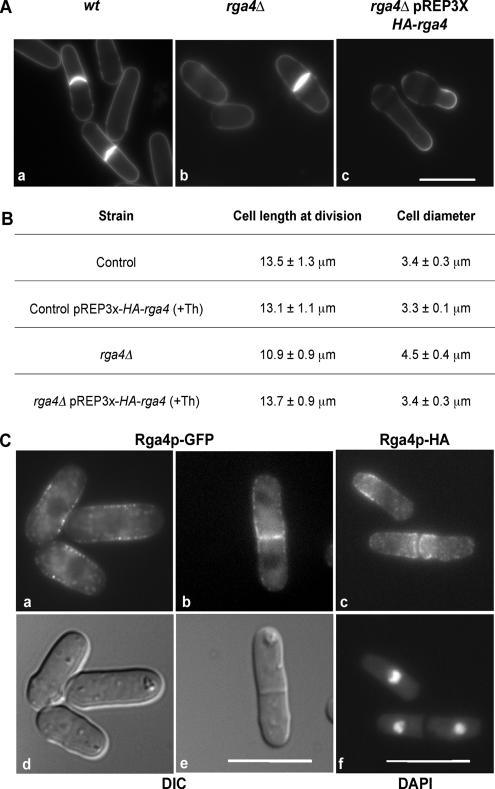
When we carefully analyzed the rga4Δ mutant phenotype, we found that loss of Rga4p results in regular shaped cells (Figure 1Ab) that are 20% shorter at time of cell division (p < 0.01; n = 25) and 33% wider (p < 0.01; n = 25) than wild-type cells (Figure 1, Aa and B). This phenotype is completely reversed to normal when a HA-tagged Rga4p protein is expressed in rga4Δ mutants at levels close to endogenous, under the control of the nmt1 promoter, in the presence of thiamine (Figure 1B; see Material and Methods for information on expression levels).
Figure 1.
Loss of Rga4p alters cellular dimensions. (A) a, wild-type 972. b, rga4Δ (FV513). c, rga4Δ strain expressing HA-Rga4p under the control of the nmt1 promoter (FV531), grown in the absence of thiamine for 16 h at 32°C. Cells were stained with calcofluor. Bar, 10 μm. (B) Effect of rga4 deletion on cell length and cell diameter at division expressed as average ± SD. Twenty-five cells were measured for each condition. (C) Localization of chromosomally tagged Rga4-GFP (FV781) and Rga4-HA (FV532). a and b, Rga4-GFP; d and e, corresponding DIC images. c, Rga4-HA; f, corresponding DAPI visualization. Note that in d and e overall cell dimensions are altered by methanol fixation. Bar, 10 μm.
In contrast, Rga4p overexpression in rga4Δ mutants, from a multicopy plasmid under the control of the nmt1 promoter, in the absence of thiamine (20 folds overexpression over endogenous levels) leads to a change in cell shape and a decrease in diameter below wild-type levels (Figure 1Ac). Under these conditions, 35% of cells display a reduced diameter (between 2 and 2.8 μm). A range of cell diameters in these cells is expected, because Rga4 is overexpressed from a multicopy Rep3X plasmid, and some plasmid instability and copy-number variability is to be anticipated. Indeed, of 96 rgaΔ cells overexpressing Rga4, 79 showed a reduction of cell diameter (compared with rga4Δ cells carrying a control plasmid), whereas 17 did not show a diameter reduction. If we consider only the 79 cells showing a reduction in diameter (which we set as a diameter value of <4 μm), we obtain an average diameter of 2.87 ± 0.4 μm, which shows a significant difference to the wild-type value of 3.41 ± 0.3 μm (n = 50) with a 99.9% confidence level (p < 0.001). If we consider all measured cells (n = 96), we obtain an average diameter of 3.14 ± 0.7 μm, which still shows a significant difference from the wild-type value, with a 98.9% confidence level. Collectively, these findings suggest that Rga4p has a role in the control of cell diameter and overall cellular dimensions in fission yeast.
Rga4 Localizes to the Cell Sides and to the Cell Septum
The Rga4p protein, when expressed as a C-terminal GFP fusion, under the control of the endogenous promoter at the rga4 locus, is found in small cytoplasmic dots that concentrate predominantly at the cell sides and at the site of cell division in live cells (Figure 1C, a and b). Furthermore, the Rga4p-GFP protein does not concentrate at the growing cell tips (Figure 1Ca; see also below in Figure 7). As a comparison, the localization of Rga4p-HA expressed under the control of its endogenous promoter at the rga4 locus was also analyzed in methanol-fixed cells (Figure 1Cc). The localization of Rga4p-HA to the cell sides and cell septum in fixed cells is very similar to the localization of Rga4p-GFP in live cell. Rga4p-HA also localizes to the cell sides and is excluded from the cell tips, although it appears to form a “collar” closer to the cell ends, rather than a more extensive “corset” pattern along the cell sides as seen with Rga4p-GFP. This difference could be due to limitations of the fixation/immunofluorescence technique used or to a slightly different stability of the Rga4p-GFP and Rga4p-HA proteins. Collectively however, these results clearly show that Rga4p is localized in a unique pattern to the cell sides and septum, and is excluded from the cell tips.
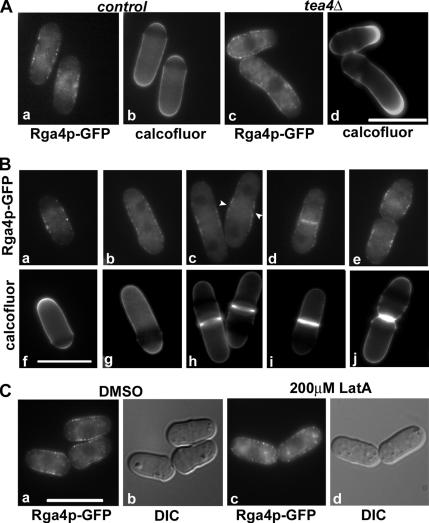
Figure 7.
Rga4p localization during cell growth. (A) a and b, control cells. c and d, tea4Δ; a and c, Rga4-GFP visualization in live cells; b and d, corresponding Calcofluor staining. (B) Localization of Rga4-GFP in cells at different stages of cell cycle. a–e, Rga4-GFP; f–j, Calcofluor staining. a and f, cell in G2 phase; b and g, cell in early mitosis, c and h, cell in mitosis during septum formation, with arrows showing the initial localization of Rga4p-GFP to the site of septation, d and i, cell during mitosis after completion of septation; e and j, cells immediately after cell separation. (C) Effect of latrunculin A on Rga4-GFP localization; a and b, DMSO-treated cells; c and d, cells treated with 200 μM latrunculin A for 20 min; a and c, Rga4-GFP visualization in live cells; b and d, DIC visualization.
The Actin Cytoskeleton Is Altered in rga4Δ Cells
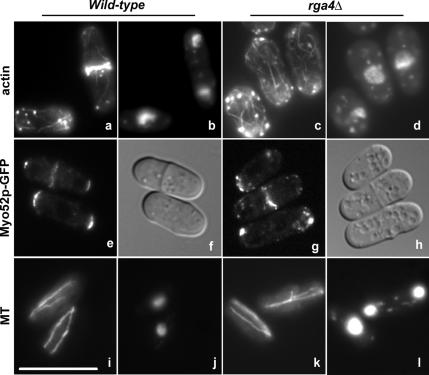
When we analyzed the actin and microtubule cytoskeletons in rga4Δ mutant cells, we found that actin patches are less concentrated at the cell tips and actin cable distribution is disorganized, with cables often polymerizing from all directions in the cell. (Figure 2, a and b). To strengthen this observation, we examined the localization of the barbed-end–directed myosin Myo52p. We found that Myo52-GFP localization is abnormal in rga4Δ cells, and is localized in more dispersed or clumped dots (Figure 2g), compared with the control cells (Figure 2e). Microtubule organization is still normal, with microtubules extending along the main cell axis (Figure 2, j and k).
Figure 2.
rga4 deletion results in disorganized cytoplasmic actin cables. a and c, phalloidin staining of actin; e and g, Myo52-GFP visualization in live cells; i and k, staining of microtubules. b, d, j, and l, DAPI staining to visualize the nucleus; f and h, DIC images; a, b, i, and j, wild-type 972; c, d, k, and l, rga4Δ (FV513); e and f, control Myo52-GFP (DM3415); g and h, rga4Δ Myo52-GFP (FV797). Cells were grown exponentially at 32°C for 24 h and then stained with phalloidin-conjugated Alexa Fluor 488 for actin and immunofluorescent anti-TAT1 staining for microtubules. Bar, 10 μm.
Loss of Rga4 Results in Asymmetric Cell Growth
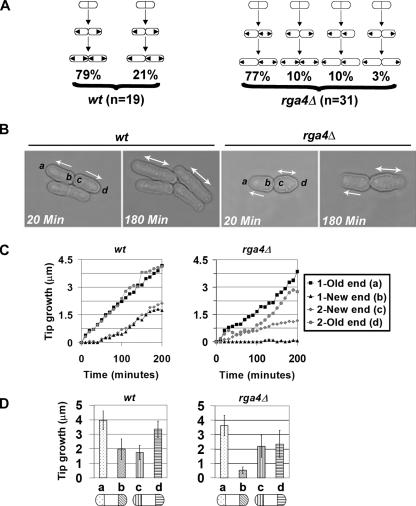
We next examined whether the normal pattern of cell growth is altered in rga4Δ mutants by performing time-lapse microscopy experiments (Figure 3, B and C). After cell division, the majority of wild-type cells (79%) grow from the tips that already existed before mitosis, generally defined as the “old ends” (Figure 3, A and B). Daughter cells continue to grow until they reach a minimal size (Mitchinson and Nurse, 1985) and then activate the new ends formed by cell division, switching from monopolar growth to bipolar growth (Figure 3A; see Supplementary Movie 1). We followed 19 pairs of wild-type daughter cells and 31 pairs of rga4Δ daughter cells for a whole cell cycle, from the initial division of their “mother” cell to the following division. We found that in 77% of rga4Δ mutant cells this overall pattern of growth is altered, so that the two daughter cells generally display an asymmetrical behavior of growth (Figure 3, A and B). After septation, one daughter cell grows from its old tip in a monopolar manner, failing to activate the new end, whereas the other daughter cell activates bipolar growth (Figure 3B; Supplementary Movie 2). Monopolar and bipolar cells were similar in overall shape, with bipolar cells displaying a slight (6%) increase in cell diameter.
Figure 3.
Pattern of polarized cell growth in rga4Δ cells. (A) Growth patterns of wild-type 972 and rga4Δ (FV513) cells and percentage of cell in each class. Arrows indicate the direction of growth in daughter cell pairs. (B) Time-lapse images of wild-type and rga4Δ daughter cell pairs. Cells were recorded every 10 min for 6 h at 25°C. (C) Example of measurement of growth at cell tips of daughter cell pairs from wild-type and rga4Δ. (Time 0 is at the time of cell separation.) (D) Total growth for each cell tip during the cell cycle for wild-type (n = 15) and in rga4Δ (n = 24) daughter cell pairs (average ± SD). Tip a is defined as the tip showing the most growth, which in rga4Δ strains is always found in the cell that does not activate the new end. Cells were grown on YE at 23–25°C (room temperature).
To quantify these differences, we measured the increase in distance from the old and new cell tips to the birth scar, in both daughter cells. In each pair of daughter cells we defined “a” as the old end that grows the most. The other ends were labeled “b,” “c,” and “d” with respect to the a end (Figure 3B). Time 0 was set at the moment when cell separation was initiated. As a typical example, the measured growth of two wild-type daughter cells and two rga4Δ daughter cells is shown in Figure 3C. This example shows that the old tips of the two wild-type cells (a and d) grow at a similar speed. Growth at the wild-type new ends (b and c) starts later and proceeds slower. In contrast, the old and new ends in the rga4Δ daughter cells each grow in a different way. The old end in the rga4 daughter cell that does not activate NETO grows the most (and it is labeled a). The b end in the same cell is inactive (Figure 3C). The new and old ends in the other daughter cell (c and d) can display variable speed of growth. After analysis of all daughter cell pairs, we found that on average the activation of the c ends occur 20 min earlier than wild type.
When we plotted the cumulative amount of growth observed in the a, b, c, and d ends (Figure 3D), we found a statistically significant difference in the growth observed at the new ends (b) and (c) in rga4Δ cells (p < 0.01; Figure 3D). These observations suggest that loss of Rga4p disrupts the symmetry of the two daughter cells and alters the time of activation of both new ends.
Loss of Rga4p Results in the Alteration of For3p Localization
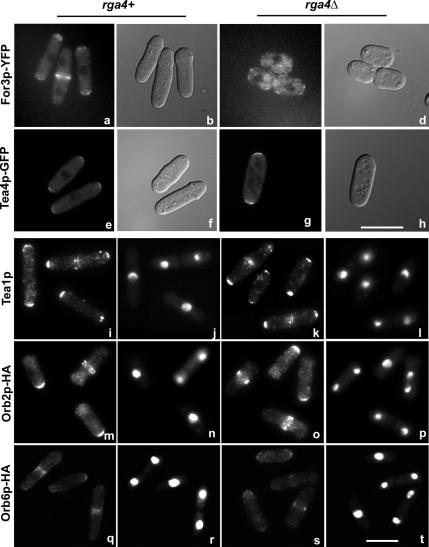
The pattern of growth observed in rga4Δ cells is similar to the one described in for3Δ mutant cells (Feierbach and Chang, 2001; Nakano et al., 2002; Sawin, 2002). For3p is a formin homology protein that has been implicated in cytoplasmic actin cable formation and microtubule organization. To determine if the localization of For3p is altered in the absence of Rga4p, we examined the localization of For3p-YFP in live rga4Δ cells (Figure 4). In the presence of Rga4p, For3p-YFP is localized predominantly to the cell tips during interphase, or to the cell septum during mitosis (Figure 4, a and b; Feierbach and Chang, 1001; Martin and Chang, 2006). In rga4Δ cells the preferential For3p-YFP localization to the cell tips is not observed, with For3p-YFP dots found localized throughout the cell cortex (Figure 4, c and d).
Figure 4.
Intracellular localization of For3p-YFP is altered in rga4 deletion cells. a, b, e, f, i, j, m, n, q, and r, control rga4+ cells; c, d, g, h, k, l, o, p, s, and t, rga4Δ cells. For3p-YFP visualization in live cells: a, BFY81; c, FV572. Tea4-GFP visualization in live cells: e, CA2301; g, FV796. DIC visualization of cells: b, d, f, and h. Tea1p visualization with a polyclonal anti-Tea1p antibody: i, wild-type, 972; k, FV513 (Mata and Nurse, 1997). Orb2p-HA visualization with an anti-HA antibody: m, FV607; o, FV610. Orb6p-HAp visualization with an anti- HA antibody: q, FV542; s, FV527. DAPI visualization of the nucleus: j, l, n, p, r, and t. Note that panels i–t are merged images of different microscopic fields. Bar, 10 μm.
To ascertain the specificity of the effect of rga4Δ on the localization of For3p, we analyzed the localization of other morphology control factors, namely Tea4p/Wsh3p, Tea1p, Orb2p/Pak1p/Shk1p protein kinase, and Orb6p protein kinase in rga4Δ cells. Tea4p/Wsh3p functions as a link between For3p and polarity marker Tea1p (Martin et al., 2005). Tea1p is a microtubule plus end factor that marks the sites of polarized cell growth (Snell and Nurse, 1994; Mata and Nurse, 1997; Behrens and Nurse, 2002; Kim et al., 2003). Orb2p/Pak1p/Shk1p is a Rho-GTPase dependent serine/threonine kinase that controls polarized cell growth and the actin cytoskeleton (Marcus et al., 1995; Ottilie et al., 1995; Verde et al., 1998). Orb6p is a ser/thr kinase with a role in cell morphogenesis and cell cycle control (Verde et al., 1998). Tea4p/Wsh3p normally localizes to both tips in control (Figure 4, e and f) and rga4Δ cells (Figure 4, g and h). Furthermore, we found that the absence of Rga4p does not affect the localization of Tea1p (Figure 4, i–l), Orb2p/Pak1p/Shk1p-HA (Figure 4, m–p), or Orb6p-HA (Figure 4, q–t), indicating that other morphological factors are not affected by loss of Rga4p.
The GAP Domain of Rga4p Is Required for Cell Diameter Control and For3p-YFP Localization
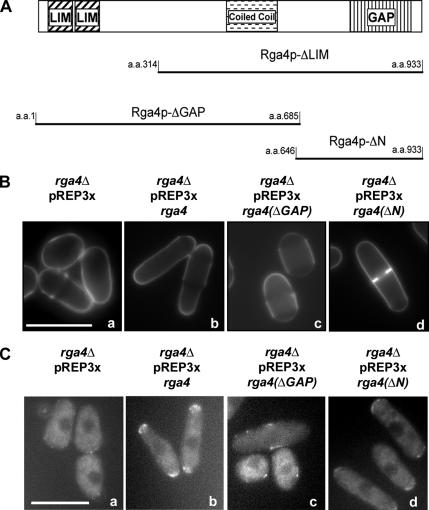
Rga4p is comprised of two N-terminal LIM domains, a coiled-coil domain, and a C-terminal GAP domain (Figure 5A). To determine the roles of these domains on the localization of For3p-YFP, we created an rga4 construct that lacks the sequence for the GAP domain (rga4-ΔGAP), a construct that lacks the sequence for the LIM domains (rga4-ΔLIM), and a construct that lacks the sequence for the LIM domains and the coiled-coil domain (rga4-ΔN), all carrying the sequence for a N-terminal HA tag. The expression level of these different fusion peptides in rga4Δ cells was similar (Das, unpublished data). We found that expression of the Rga4p GAP domain, at levels close to endogenous, is able to suppress the cell diameter defect of rga4Δ mutants. rga4Δ cells expressing the GAP domain alone (HA-Rga4p-ΔN; Figure 5Bd) displayed a cell diameter (4.0 ± 0.2 μm) that is significantly narrower than what observed in control rga4Δ cells (4.9 ± 0.3 μm; p < 0.001; Figure 5Ba). Conversely, expression of the N-terminal domain (HA-Rga4p-ΔGAP) in rga4Δ cells does not rescue the cell diameter defect of rga4Δ cells (Figure 5Bc).
Figure 5.
The GAP domain of Rga4p is required for For3p-YFP localization and for regulation of cell diameter. (A) LIM domains, coiled-coil domain and GAP domain in the Rga4p protein and the different constructs used in this study. (B) Calcofluor staining of rga4Δ cells expressing different domains of Rga4p; a, rga4Δ cells with pREP3X (FV516); b, rga4Δ cells expressing HA-Rga4p (FV531); c, rga4Δ cells expressing HA-Rga4p-ΔGAP (FV575); d, rga4Δ cells expressing HA-Rga4p-ΔN (FV574). Cells were grown for eight generations in the presence of thiamine. (C) For3p-YFP localization in rga4Δ cells expressing different domains of Rga4p. a, rga4Δ cells carrying empty pREP3X (FV516); b, rga4Δ cells expressing full-length HA-Rga4p (FV516); c, rga4Δ cells expressing HA-Rga4p-ΔGAP (FV575); d, rga4Δ cells expressing HA-Rga4p-ΔN (FV574). Cells were grown for 20 h at 25°C in the absence of thiamine. Bar, 10 μm.
Furthermore, we found that the expression of the GAP domain of Rga4 (HA-Rga4p-ΔN: Figure 5Cd) in rga4Δ cells is sufficient to restore the localization of For3p-YFP to the cell tip(s) and the cell septum. However, although low levels of expression of the full-length Rga4 is able to restore For3p localization in rga4Δ cells, expression of the GAP domain at levels close to endogenous is not sufficient to restore For3p localization. Expression of the GAP domain restores For3p localization only when expressed at levels sixfold higher than endogenous (see Material and Methods for details on expression conditions and Supplementary Figure 1A for a quantification of For3p localization). This is likely due to the fact that in the absence of the N-terminus, Rga4 GAP domain does not localize efficiently to the cell cortex (see Supplementary Figure 1B). At these levels of Rga4 expression the actin cytoskeleton was not visibly altered either in wild type or in rga4Δ cells expressing For3p-YFP, as shown in Supplementary Figure 2.
Collectively, our results suggest that the C-terminal region of Rga4p (the GAP domain) is required for cell diameter control and for proper localization of For3p-YFP.
Overexpression of the N-terminus of Rga4p Induces For3p-dependent Actin Cable Formation
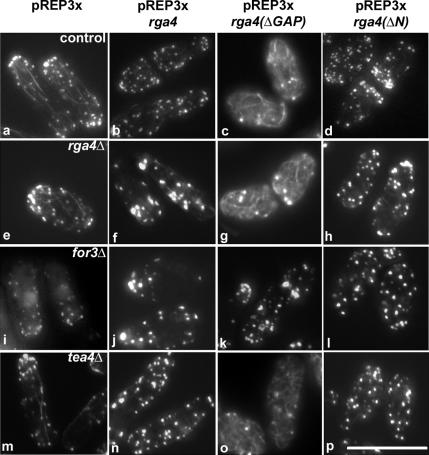
Because the formin For3p functions in the control of actin cable polymerization, we tested the effects of overexpressing HA-Rga4p-ΔGAP and HA-Rga4p-ΔN (20-fold more than the normal level) on the actin cytoskeleton (Figure 6). We found that overexpression of the N-terminal domain of Rga4p (containing the LIM domains and the coil-coiled domain) substantially increases the number of actin cables found in wild-type (Figure 6c) and rga4Δ cells (Figure 6g). Overexpression of full-length HA-Rga4p or the GAP domain alone (HA-Rga4p-ΔN) at comparable levels does not induce actin cable formation in control or rga4Δ cells (Figure 6, b and d, f and h), although it resulted in the loss of polarized localization of actin patches.
Figure 6.
Overexpression of the N-terminus of Rga4p induces actin cable formation in a For3p-dependent manner. (a) Wild-type cells carrying an empty Rep3X plasmid (FV800). (b) Wild-type cells expressing full length HA-Rga4p (FV801). (c) Wild-type cells expressing HA-Rga4p-ΔGAP (FV804). (d) Wild-type cells expressing HA-Rga4p- ΔN (FV802). (e) rga4Δ cells carrying an empty Rep3X plasmid (FV416). (f) rga4Δ cells expressing full length HA-Rga4p (FV531). (g) rga4Δ cells expressing HA-Rga4p-ΔGAP (FV575). (h) rga4Δ cells expressing HA-Rga4p- ΔN (FV574). (i) for3Δ cells carrying an empty Rep3X plasmid (FV605). (j) for3Δ cells expressing HA-Rga4p (FV606). (k) for3Δ cells expressing HARga4p- ΔGAP (FV604). (l) for3Δ cells expressing HA-Rga4p-ΔN (FV603). (m) tea4Δ cells carrying an empty Rep3X plasmid (FV759). (n) tea4Δ cells expressing HA-Rga4p (FV760). (o) tea4Δ cells expressing HA-Rga4p-ΔGAP (FV761). (p) tea4Δ cells expressing HA-Rga4p-ΔN (FV762). Cells were grown for 16 h at 32°C in the absence of thiamine. Bar, 10 μm.
Because the function of For3p is to promote actin cable polymerization, we tested if the effects induced by HA-Rga4p-ΔGAP expression are dependent on For3p. The induction of actin cable formation by expression of the N-terminus of Rga4p (HA-Rga4-ΔGAP) was found to require the presence of For3p, because no actin cable formation was observed when HA-Rga4p-ΔGAP is overexpressed in the for3Δ mutant (Figure 6k).
The For3p-interacting protein Tea4p was also previously shown to promote actin cable formation when overexpressed (Martin et al., 2005). However, we found that the induction of actin cable formation by expression of the N-terminus of Rga4p (HA-Rga4-ΔGAP) is not dependent on the presence of Tea4p (Figure 6o). These results indicate that the N-terminus of Rga4p promotes actin cable formation when overexpressed, a function that is functionally dependent on the formin For3p, but not Tea4p/Wsh3p.
Control of Rga4p Localization during Cell Growth
Because we found that Rga4p-GFP localizes predominantly to the cell sides and not at the cell tips during cell growth, we explored how this localization correlates with tip growth, by visualizing Rga4p-GFP in bipolar wild-type cells (Figure 7A, a and b) and monopolar tea4Δ cells (Figure 7A, c and d), and by staining the cell wall with Calcofluor. We found that a decrease of Rga4p-GFP protein concentration strongly correlates with initiation of tip growth, and that in monopolar tea4Δ cells, Rga4p-GFP concentration only decreases at the growing tip, not at the nongrowing one (Figure 7A, c and d). We also confirmed these observations by time-lapse microscopy in wild-type and tea4Δ cells: in tea4Δ cells, where growth is asymmetrical in the two daughter cells, Rga4p-GFP concentration decreases only at the growing tips, which are the old end in one daughter cell and the new end in the other daughter cell (data not shown).
We also examined Rga4p-GFP localization during cell division (Figure 7B). We analyzed 100 cells at various stages of septum assembly, as determined by Calcofluor staining. We found that Rga4p-GFP localizes at the cell division site only after the initiation of septum formation (Figure 7B, c, d, h, and i). Rga4p-GFP localization at the cell septum is lost following cell separation (Figure 7B, e and j).
Finally, we tested if Rga4p localization is dependent on actin. As shown in Figure 7Cc, treatment with 200 μM Latrunculin A for up to 1 h did not alter Rga4p-GFP localization to the cell sides, although it was sufficient to depolymerize actin (not shown; Feierbach and Chang, 2001). However, in latrunculin-treated cells Rga4p-GFP was found to localize closer to and sometimes all the way up to the cell tips in 93% (n = 30) of the cells, compared with the control DMSO-treated cells where only 7% (n = 30) of the cells showed some tip localization of Rga4p-GFP. Thus, it is possible that the exclusion of Rga4p-GFP from the cell tips is an actin- and growth-dependent process. In conclusion, our observations indicate that Rga4-GFP localization at the cell tips inversely correlates with cell tip growth.
DISCUSSION
Rga4p Controls Cellular Dimensions and Cell Diameter
In this article we examine the function of Rga4p in fission yeast S. pombe cell morphogenesis. Rga4p is a predicted GTPase-activator protein for Rho-like GTPases and is similar to Rga1p and Rga2p from S. cerevisiae (Stevenson et al., 1995; Nakano et al., 2001; Smith et al., 2002). rga4 is nonessential for cell viability (Nakano et al., 2001; our unpublished results). In this article, we show that in the absence of Rga4p, cellular dimensions are altered, as cells become 33% wider in cell diameter. This effect is not due to a nonspecific disruption of cell morphology, because cell dimensions in rga4Δ mutant cells are regular and remarkably homogenous, with individual values narrowly deviating from the average. Maintenance of overall cell shape is also observed in rga4Δ cdc10-129 mutant cells cultured at 35.5°C, which continue to grow in a polarized manner (Das, unpublished data). Furthermore, cell growth in rga4Δ cells proceeds well, with rates similar to wild-type cells. In rga4Δ cells cell length at division is decreased and overall cell volume (calculated approximating the S. pombe cell to a cylinder) is increased by 40%. This rga4Δ phenotype is novel, because cell diameter mutants have not been previously described in fission yeast. Generally, mutants in proteins involved in the control of cell morphology display a more global alteration of cell polarity and shape, making the study of mechanisms controlling cell dimensions difficult.
How does Rga4p control cell diameter? One possibility is that Rga4p limits the size or the activity of the zone of polarized cell growth at the cell tip. This interpretation is suggested by three observations. First, the Rga4p protein is intriguingly localized in a collar or corset-like pattern that extends along the nongrowing (lateral) areas of the cell up to the interface with the growing tip. Second, loss of Rga4p affects the width of the cell but does not interfere with the ability of the cell to assemble an actin ring or a septum and to divide. Third, overexpression of Rga4p leads to a narrowing of the areas of polarized cell growth with a resulting discontinuity in the shape of the cell tips. Thus, these observations suggest that cell diameter can be regulated, and that Rga4p may function by spatially restricting cell growth at the cell tips.
Our study suggests that expression of the C-terminal GAP domain is able to restore control of cell diameter in rga4Δ cells. Our laboratory is currently investigating the nature of the GTPase that is targeted by Rga4p. Interestingly, the related S. cerevisiae proteins Rga1p and Rga2p were shown to have a role in mediating the interaction of Cdc42p GTPase with the PAK-like kinase Ste20p and to facilitate the role of Cdc42p in septin organization (Smith et al., 2002; Caviston et al., 2003; Longtine and Bi, 2003). Our own data indicate that Rga4p has a role in formin For3p localization and actin cable polymerization (discussed below). For3p has been shown to interact with GTPases Cdc42p and Rho3p (Nakano et al., 2002). Thus, it is likely that Rga4p may control the activity of several GTPase-mediated cellular functions at the cell tips.
Rga4p Functions in a Pathway Controlling Formin For3p Localization and Actin Cable Formation
rga4Δ cells also display an altered localization of actin cables and patches. In rga4Δ cells, actin cables are also more readily visible and appear thicker and more numerous, although the actual quantification of actin cable number is difficult. Consistent with a partial disruption of the normal actin cytoskeleton, the localization of the barbed-end directed myosin Myo52-GFP is also altered.
Several lines of evidence indicate that Rga4p functionally interacts with the formin homology protein For3p. First, we found that rga4Δ mutants and for3Δ mutants display the same alterations in the establishment and spatial control of polarized cell growth in daughter cells (this article, see above; Feierbach and Chang, 2001). Second, For3p depends on Rga4p for normal localization to the cell tips. When Rga4p is absent, For3p dots are found in a more dispersed pattern throughout the cell cortex, including the side of the cell. This phenotype can be rescued by expression of the GAP domain alone. A disturbance in the polarized localization of For3p in the rga4Δ mutant could explain why in these cells actin cables appear more disorganized. Third, overexpression of the N-terminus of Rga4p strongly induces actin cable formation in wild-type and rga4Δ mutants but not in for3Δ mutants, suggesting that Rga4p-ΔGAP–dependent promotion of actin polymerization is For3p dependent. This strong promotion of actin cable formation when the Rga4p GAP domain is deleted could be the result of interaction with other proteins, leading to the stabilization of a promoter of actin cable polymerization or sequestration of an inhibitor of actin cable polymerization. However, it is not due to increased For3p protein levels, because we found they were unchanged by Rga4p overexpression (Das, unpublished data). Collectively, our observations suggest that Rga4p and For3p function in a common pathway controlling actin cable formation and that Rga4p functions to restrict For3p localization to the correct sites of polarized cell growth.
Rga4p Has Different Roles in the Control of Cell Growth
The role of Rga4p in the establishment of cell diameter and in the control of For3p localization is possibly exerted by establishing a “no growth” domain at the cell sides, where Rga4p is localized as a “corset” that extends along the cell length. At the present time, the Rga4p pattern of localization is novel and unique, at least for proteins involved in the control of cell morphology. Several factors have been described that localize in a centrally located cortical band overlying the nucleus during interphase, like Cdr2p, a protein involved in nutritional sensing of cell size control during mitosis (Kanoh and Russell, 1998; Morrell et al., 2004) or Mid1 (Sohrmann et al., 1996; Bahler et al., 1998; Paoletti and Chang, 2000; Wu et al., 2006); however, these bands do not extend all along the cell sides and are narrower in width. Furthermore, these proteins reorganize in a ring at the time of actin ring formation, whereas Rga4p does not localize to the site of cell division until the septum is formed, and maintains its localization at the cell sides. Thus it is likely that the lateral cell cortex is able to define several domains that act as independent positional markers, regulating different morphological functions.
Interestingly, Rga4p also localizes to nongrowing cell tips, suggesting that Rga4p defines cell ends that are not competent for growth. Rga4p has a function in maintenance of daughter cell symmetry, because in rga4Δ mutants one daughter cell fails to activate NETO, whereas the other activates both tips almost simultaneously. It should be noted that the rga4Δ pattern of growth is sensitive to the temperature of growth and is observed at 23–25°C, which is the temperature at which time-lapse experiments were performed. When we increased the temperature to 32°C, we noticed that at this temperature both daughter cells often activated both tips simultaneously. Thus, these observations suggest the asymmetrical pattern of growth in rga4Δ daughter cells could be the product of two processes: a relief of cell growth inhibition at the new end in both daughter cells, independent of temperature, and a decreased ability of one of the two new ends to activate cell growth, which can be overcome by increasing temperature. Thus it is likely that at least one of the roles of Rga4p at the site of cell division is a negative regulation of cell growth, preventing precocious activation of the new end. Interestingly, Rga4p localizes at the site of cell division after the initiation of septum formation and is lost from this site following cell separation. It is possible that this transient localization of Rga4p has a role in preventing abnormal new end activation after mitosis. A pattern of growth similar to the one of rga4Δ mutants has been previously described in for3Δ mutants (Feierbach and Chang, 2001). In rga4 mutants, however, the For3p protein is still able to promote actin cable formation, and is still present at the cell cortex, although it is much less concentrated at the cell tips. Thus, the alteration of growth control observed in the rga4Δ phenotype suggests that, if the function of For3p is required for symmetry, a critical threshold of For3p concentration at the cell tips is important for normal pattern of cell growth. Alternatively, daughter cell asymmetry is the result of a more extensive change in the structure or protein composition of large multifunctional protein complexes that regulate cell polarity at the cell tips. Indeed, For3p localization to one of the two cell tips is regulated by interaction with the Tea4p/Whs3p protein complex (Martin et al., 2005). However, For3p is also thought to bind to other, yet unidentified factors, which allow its localization to the second tip (Martin et al., 2005). Thus, it is possible that loss of Rga4p also has a role in the localization of other morphological factors with a role in the control of cell symmetry.
Collectively, our findings indicate that Rga4p plays a role in fine tuning several aspects of cell morphogenesis and provide an initial insight into a molecular mechanism of cell diameter control. The future analysis of the proteins interacting with Rga4p and controlling its activity will likely increase our understanding of complex morphogenesis control mechanisms.
Supplementary Material
ACKNOWLEDGMENTS
We thank Dr. Paul Nurse, Dr. Fred Chang, Dr. Kazuhiro Shiozaki, Dr. Juan Mata, Dr. Dannel McCollum and Dr. Keith Gull for the various strains and antibodies. We thank Dr. Sandra Lemmon and Dr. Gennaro D'Urso for critically reading the manuscript, and the Yeast Club at the University of Miami for useful suggestions. This work was supported by a National Science Foundation Grant NSF 0344798 and by the Sylvester Cancer Center, University of Miami Miller School of Medicine.
Abbreviations used:
- GAP
GTPase-activating protein.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E06-09-0883) on March 21, 2007.

 The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
The online version of this article contains supplemental material at MBC Online (http://www.molbiolcell.org).
REFERENCES
- Arellano M., Duran A., Perez P. Rho 1 GTPase activates the (1–3)beta D-glucan synthase and is involved in Schizosaccharomyces pombe morphogenesis. EMBO J. 1996;15(17):4584–4591. [PMC free article] [PubMed] [Google Scholar]
- Bahler J., Steever A. B., Wheatley S., Wang Y., Pringle J. R., Gould K. L., McCollum D. Role of polo kinase and Mid1p in determining the site of cell division in fission yeast. J. Cell Biol. 1998;143(6):1603–1616. doi: 10.1083/jcb.143.6.1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardin A. J., Le Borgne R., Schweisguth F. Asymmetric localization and function of cell-fate determinants: a fly's view. Curr. Opin. Neurobiol. 2004;14(1):6–14. doi: 10.1016/j.conb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- Behrens R., Nurse P. Roles of fission yeast tea1p in localization of polarity factors and in organizing the microtubular cytoskeleton. J. Cell Biol. 2002;157(5):783–793. doi: 10.1083/jcb.200112027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calonge T. M., Arellano M., Coll P. M., Perez P. Rga5p is a specific Rho1p GTPase-activating protein that regulates cell integrity in Schizosaccharomyces pombe. Mol. Microbiol. 2003;47(2):507–518. doi: 10.1046/j.1365-2958.2003.03312.x. [DOI] [PubMed] [Google Scholar]
- Caviston J. P., Longtine M., Pringle J. R., Bi E. The role of Cdc42p GTPase-activating proteins in assembly of the septin ring in yeast. Mol. Biol. Cell. 2003;14(10):4051–4066. doi: 10.1091/mbc.E03-04-0247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang F., Peter M. Yeasts make their mark. Nat. Cell Biol. 2003;5(4):294–299. doi: 10.1038/ncb0403-294. [DOI] [PubMed] [Google Scholar]
- Chang F., Verde F. Molecular Biology of Schizosaccharomyces pombe. Springer-Verlag; 2004. Control of cell polarity and cell morphogenesis; pp. 255–267. [Google Scholar]
- Chang F., Woolard A., Nurse P. Isolation and characterization of fission yeast mutants defective in the assembly and placement of the contractile actin ring. J. Cell Sci. 1996;109:131–142. doi: 10.1242/jcs.109.1.131. [DOI] [PubMed] [Google Scholar]
- Chen G. C., Zheng L., Chan C. S. The LIM domain-containing Dbm1 GTPase-activating protein is required for normal cellular morphogenesis in Saccharomyces cerevisiae. Mol. Cell. Biol. 1996;16(4):1376–1390. doi: 10.1128/mcb.16.4.1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faix J., Grosse R. Staying in shape with formins. Dev. Cell. 2006;10(6):693–706. doi: 10.1016/j.devcel.2006.05.001. [DOI] [PubMed] [Google Scholar]
- Feierbach B., Chang F. Roles of the fission yeast formin for3p in cell polarity, actin cable formation and symmetric cell division. Curr. Biol. 2001;11:1656–1665. doi: 10.1016/s0960-9822(01)00525-5. [DOI] [PubMed] [Google Scholar]
- Galway M. E., Lane D. C., Schiefelbein J. W. Defective control of growth rate and cell diameter in tip-growing root hairs of the rhd4 mutant of Arabidopsis thaliana. Can. J. Bot. 1999;77:494–507. [Google Scholar]
- Higgs H. N. Formin proteins: a domain-based approach. Trends Biochem. Sci. 2005;30(6):342–353. doi: 10.1016/j.tibs.2005.04.014. [DOI] [PubMed] [Google Scholar]
- Johnson B. F., Lu C. Morphometric analysis of yeast cells. IV. Increase of the cylindrical diameter of Schizosaccharomyces pombe during the cell cycle. Exp. Cell Res. 1975;95(1):154–158. doi: 10.1016/0014-4827(75)90620-5. [DOI] [PubMed] [Google Scholar]
- Kanoh J., Russell P. The protein kinase Cdr2, related to Nim1/Cdr1 mitotic inducer, regulates the onset of mitosis in fission yeast. Mol. Biol. Cell. 1998;9(12):3321–3334. doi: 10.1091/mbc.9.12.3321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H., Yang P., Catanuto P., Verde F., Lai H., Du H., Chang F., Marcus S. The kelch repeat protein, Tea1, is a potential substrate target of the p21-activated kinase, Shk1, in fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2003;278(32):30074–30082. doi: 10.1074/jbc.M302609200. [DOI] [PubMed] [Google Scholar]
- Kovar D. R. Molecular details of formin-mediated actin assembly. Curr. Opin. Cell Biol. 2006;18(1):11–17. doi: 10.1016/j.ceb.2005.12.011. [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Harris E. S., Mahaffy R., Higgs H. N., Pollard T. D. Control of the assembly of ATP- and ADP-actin by formins and profilin. Cell. 2006;124(2):423–435. doi: 10.1016/j.cell.2005.11.038. [DOI] [PubMed] [Google Scholar]
- Kovar D. R., Kuhn J. R., Tichy A. L., Pollard T. D. The fission yeast cytokinesis formin Cdc12p is a barbed end actin filament capping protein gated by profilin. J. Cell Biol. 2003;161(5):875–887. doi: 10.1083/jcb.200211078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubitschek H. E., Clay K. B. A second growth state for Schizosaccharomyces pombe. Exp. Cell Res. 1986;165(1):243–54. doi: 10.1016/0014-4827(86)90548-3. [DOI] [PubMed] [Google Scholar]
- Longtine M. S., Bi E. Regulation of septin organization and function in yeast. Trends Cell Biol. 2003;13(8):403–409. doi: 10.1016/s0962-8924(03)00151-x. [DOI] [PubMed] [Google Scholar]
- Marcus S., Polverino A., Chang E., Robbins D., Cobb M. H., Wigler M. H. Shk1, a homolog of the Saccharomyces cerevisiae Ste20 and mammalian p65PAK protein kinases, is a component of a Ras/Cdc42 signaling module in the fission yeast Schizosaccharomyces pombe. Proc. Natl. Acad. Sci. USA. 1995;92(13):6180–6104. doi: 10.1073/pnas.92.13.6180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin S. G., Chang F. Dynamics of the formin for3p in actin cable assembly. Curr. Biol. 2006;16(12):1161–1170. doi: 10.1016/j.cub.2006.04.040. [DOI] [PubMed] [Google Scholar]
- Martin S. G., McDonald W. H., Yates J. R., 3rd, Chang F. Tea4p links microtubule plus ends with the formin for3p in the establishment of cell polarity. Dev. Cell. 2005;8(4):479–491. doi: 10.1016/j.devcel.2005.02.008. [DOI] [PubMed] [Google Scholar]
- Mata J., Nurse P. tea1 and the microtubular cytoskeleton are important for generating global spatial order within the fission yeast cell. Cell. 1997;89(6):939–949. doi: 10.1016/s0092-8674(00)80279-2. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. 1993;123(1):127–130. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- Miller P. J., Johnson D. I. Cdc42p GTPase is involved in controlling polarized cell growth in Schizosaccharomyces pombe. Mol. Cell. Biol. 1994;14(2):1075–1083. doi: 10.1128/mcb.14.2.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchinson J. M. The growth of single cells. I. Schizosaccharomyces pombe. Exp. Cell Res. 1957;13:244–262. doi: 10.1016/0014-4827(57)90005-8. [DOI] [PubMed] [Google Scholar]
- Mitchinson J. M. Physiological and cytological methods for Schizosaccharomyces pombe. In: Prescott D. M., editor. Methods of Cell Physiology. Vol. 4. 1970. pp. 131–165. Academic Press. [Google Scholar]
- Mitchison J. M., Nurse P. Growth in cell length in the fission yeast Schizosaccharomyces pombe. J. Cell Sci. 1985;75:357–376. doi: 10.1242/jcs.75.1.357. [DOI] [PubMed] [Google Scholar]
- Moreno S., Klar A., Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Morrell J. L., Nichols C. B., Gould K. L. The GIN4 family kinase, Cdr2p, acts independently of septins in fission yeast. J. Cell Sci. 2004;117(22):5293–5302. doi: 10.1242/jcs.01409. [DOI] [PubMed] [Google Scholar]
- Nakano K., Mabuchi I. Isolation and sequencing of two cDNA clones encoding Rho proteins from the fission yeast Schizosaccharomyces pombe. Gene. 1995;155(1):119–122. doi: 10.1016/0378-1119(94)00917-h. [DOI] [PubMed] [Google Scholar]
- Nakano K., Arai R., Mabuchi I. The small GTP-binding protein Rho1 is a multifunctional protein that regulates actin localization, cell polarity, and septum formation in the fission yeast Schizosaccharomyces pombe. Genes Cells. 1997;2(11):679–694. doi: 10.1046/j.1365-2443.1997.1540352.x. [DOI] [PubMed] [Google Scholar]
- Nakano K., Mutoh T., Mabuchi I. Characterization of GTPase-activating proteins for the function of the Rho-family small GTPases in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2001;6(12):1031–1042. doi: 10.1046/j.1365-2443.2001.00485.x. [DOI] [PubMed] [Google Scholar]
- Nakano K., Imai J., Arai R., Akio T., Matsui Y., Mabuchi I. The small GTPase Rho3 and the diaphanous/formin For3 function in polarized cell growth in fission yeast. J. Cell Sci. 2002;115:4629–4639. doi: 10.1242/jcs.00150. [DOI] [PubMed] [Google Scholar]
- Nelson W. J. Adaptation of core mechanisms to generate cell polarity. Nature. 2003;422(6933):766–774. doi: 10.1038/nature01602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nurse P. Genetic control of cell size at cell division in yeast. Nature. 1975;256:547–551. doi: 10.1038/256547a0. [DOI] [PubMed] [Google Scholar]
- Ottilie S., Miller P. J., Johnson D. I., Creasy C. L., Sells M. A., Bagrodia S., Forsburg S. L., Chernoff J. Fission yeast pak1+ encodes a protein kinase that interacts with Cdc42p and is involved in the control of cell polarity and mating. EMBO J. 1995;14(23):5908–5919. doi: 10.1002/j.1460-2075.1995.tb00278.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti A., Chang F. Analysis of mid1p, a protein required for placement of the cell division site, reveals a link between the nucleus and the cell surface in fission yeast. Mol. Biol. Cell. 2000;11(8):2757–2773. doi: 10.1091/mbc.11.8.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelham R. J., Chang F. Role of actin polymerization and actin cables in actin-patch movement in Schizosaccharomyces pombe. Nat. Cell Biol. 2001;3:235–244. doi: 10.1038/35060020. [DOI] [PubMed] [Google Scholar]
- Preuss M. L., Serna J., Falbel T. G., Bednarek S. Y., Nielsen E. The Arabidopsis Rab GTPase RabA4b localizes to the tips of growing root hair cells. Plant Cell. 2004;16(6):1589–1603. doi: 10.1105/tpc.021634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero S., Le Clainche C., Didry D., Egile C., Pantaloni D., Carlier M. F. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119(3):419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- Sano M., Schneider M. D. Cyclins that don't cycle—cyclin T/cyclin- dependent kinase-9 determines cardiac muscle cell size. Cell Cycle. 2003;2(2):99–104. [PubMed] [Google Scholar]
- Sawin K. E. Cell polarity: following formin function. Curr. Biol. 2002;12(1):R6–R8. doi: 10.1016/s0960-9822(01)00637-6. [DOI] [PubMed] [Google Scholar]
- Smith L. G. Cytoskeletal control of plant cell shape: getting the fine points. Curr. Opin. Plant Biol. 2003;6(1):63–73. doi: 10.1016/s1369-5266(02)00012-2. [DOI] [PubMed] [Google Scholar]
- Smith G. R., Givan S. A., Cullen P., Sprague G. F., Jr GTPase-activating proteins for Cdc42. Eukaryot. Cell. 2002;1(3):469–480. doi: 10.1128/EC.1.3.469-480.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell V., Nurse P. Genetic analysis of cell morphogenesis in fission yeast—a role for casein kinase II in the establishment of polarized growth. EMBO J. 1994;13(9):2066–2074. doi: 10.1002/j.1460-2075.1994.tb06481.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohrmann M., Fankhauser C., Brodbeck C., Simanis V. The dmf1/mid1 gene is essential for correct positioning of the division septum in fission yeast. Genes Dev. 1996;10(21):2707–2719. doi: 10.1101/gad.10.21.2707. [DOI] [PubMed] [Google Scholar]
- Stevenson B. J., Ferguson B., De Virgilio C., Bi E., Pringle J. R., Ammerer G., Sprague G. F., Jr Mutation of RGA1, which encodes a putative GTPase activating protein for the polarity-establishment protein Cdc42p, activates the pheromone-response pathway in the yeast Saccharomyces cerevisiae. Genes Dev. 1995;9(23):2949–2963. doi: 10.1101/gad.9.23.2949. [DOI] [PubMed] [Google Scholar]
- Streiblova E., Wolf A. Cell wall growth during the cell cycle of Schizosaccharomyces pombe. Z. Allg. Mikrobiol. 1972;12:673–684. [PubMed] [Google Scholar]
- Tatebe H., Shimada K., Uzawa S., Morigasaki S., Shiozaki K. Wsh3/Tea4 is a novel cell-end factor essential for bipolar distribution of Tea1 and protects cell polarity under environmental stress in S. pombe. Curr. Biol. 2005;15(11):1006–1015. doi: 10.1016/j.cub.2005.04.061. [DOI] [PubMed] [Google Scholar]
- Verde F. On growth and form: control of cell morphogenesis in fission yeast. Curr. Opin. Microbiol. 1998;1(6):712–718. doi: 10.1016/s1369-5274(98)80120-3. [DOI] [PubMed] [Google Scholar]
- Verde F., Wiley D. J., Nurse P. Fission yeast orb6, a ser/thr protein kinase related to mammalian rho kinase and myotinc dystrophy kinase, is required for maintenance of cell polarity and coordinates cell morphogenesis with the cell cycle. Proc. Natl. Acad. Sci. USA. 1998;95:7526–7531. doi: 10.1073/pnas.95.13.7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiley D. J., Marcus S., D'Urso G., Verde F. Control of cell polarity in fission yeast by association of Orb6p kinase with the highly conserved protein methyltransferase Skb1p. J. Biol. Chem. 2003;278(27):25256–25263. doi: 10.1074/jbc.M209703200. [DOI] [PubMed] [Google Scholar]
- Wu J. Q., Sirotkin V., Kovar D. R., Lord M., Beltzner C. C., Kuhn J. R., Pollard T. D. Assembly of the cytokinetic contractile ring from a broad band of nodes in fission yeast. J. Cell Biol. 2006;174(3):391–402. doi: 10.1083/jcb.200602032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P., Qyang Y., Bartholomeusz G., Zhou X., Marcus S. The novel Rho GTPase-activating protein family protein, Rga8, provides a potential link between Cdc42/p21-activated kinase and Rho signaling pathways in the fission yeast, Schizosaccharomyces pombe. J. Biol. Chem. 2003;278(49):48821–48830. doi: 10.1074/jbc.M306819200. [DOI] [PubMed] [Google Scholar]
- Zigmond S. H. Formin-induced nucleation of actin filaments. Curr. Opin. Cell Biol. 2004;16(1):99–105. doi: 10.1016/j.ceb.2003.10.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.