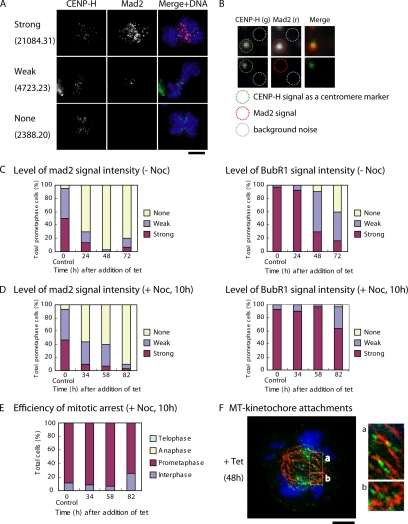
Figure 4.
Localization of mitotic checkpoint proteins in CENP-C–deficient cells. (A) Immunofluorescence analysis of Mad2 in CENP-C–deficient cells. Because cells with various intensities of Mad2 signals are observed, we classified cells into three groups on the basis of arbitrary unit values of Mad2 signal intensity: strong (unit value of signal intensity >7001), weak (unit value of signal intensity, 3001–7000), and none (unit value of signal intensity <3000). The signal intensities of Mad2 and BubR1 for >30 kinetochores in each cell were measured, and the average value per individual cell was calculated. Unit value of signal intensity per cell is shown. Typical examples for measurements are shown in B. Bars, 10 μm. (B) Examples of quantification of Mad2 level. CENP-H was used as a marker of the centromere region. Mad2 signal intensities for each kinetochore were measured, and background noise was subtracted. (C) Quantification of cells with various Mad2 and BubR1 signal intensities in CC6-121-13 cells after inhibition of CENP-C transgene expression after addition of tet at time 0. Because we confirmed that amount of CENP-C at time 0 is similar to that of wild-type cells, CC6-121-13 cells at time 0 are used as control cells. (D) Quantification of Mad2 and BubR1 signals in CC6-121-13 cells after inhibition of CENP-C transgene expression after addition of tet at time 0. In these experiments, cells were treated with nocodazole for 10 h before immunofluorescence analysis. (E) Mitotic index after treatment of CENP-C–deficient cells with the spindle poison nocodazole. (F) Immunofluorescence analysis of CC6-121-13 cells expressing CENP-H-GFP (green) with anti-α-tubulin (red) at 48 h after tet addition (+tet). Many chromosomes seem to have formed kinetochore–microtubule attachments. Bars, 5 μm.