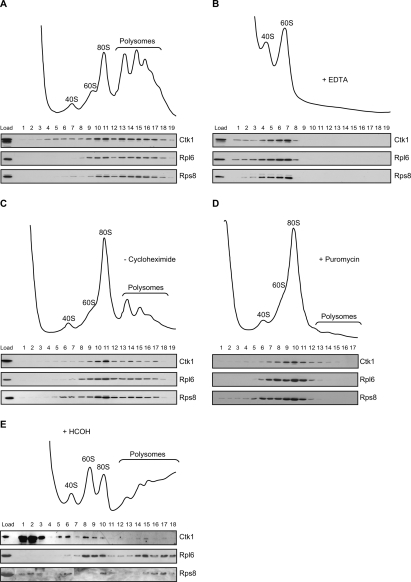
Figure 1.
Ctk1 associates with polysomes. (A) Sedimentation behavior of Ctk1-TAP on sucrose density gradients. (Top panel) Ribosomal fractions (40S, 60S, 80S, and polysomes) were determined by OD254 nm measurement of the gradient fractions. (Bottom panel) The presence of Ctk1-TAP in each fraction was visualized by Western blotting using an anti-protein A antibody. (Bottom panel) For comparison, the sedimentation behavior of a ribosomal protein of the large ribosomal subunit (Rpl6) and of the small ribosomal subunit (Rps8) is shown. (B, top panel) Sedimentation behavior of Ctk1-TAP on sucrose density gradients containing EDTA, which disrupts mono- and polysomes. (Bottom panel) Ctk1 shifts to the fractions containing ribosomal subunits. (C) The distribution of Ctk1 shifts to the 80S fraction when cycloheximide is omitted. The sucrose density gradient is as in A, but lacking cycloheximide. Without cycloheximide the ribosomes run off the mRNA, and the amount of polysomes is specifically reduced. Compared with A, Ctk1, Rpl6, and Rps8 shift to the fractions of the 80S peak. (D) Ctk1 is lost from the high-density fractions in gradients with puromycin. The sucrose density gradient is as in C, but containing puromycin, which leads to a complete loss of polysomes. Ctk1 as well as Rpl6 and Rps8 are lost from the high-density fractions. (E) Ctk1 binds to ribosomes in vivo. Cells were cross-linked with formaldehyde prior to lysis of the cells in buffer containing 500 mM KCl and separation of the lysate on a 500 mM KCl sucrose density gradient. Ctk1, Rpl6, and Rps8 were visualized by Western blotting of each fraction.