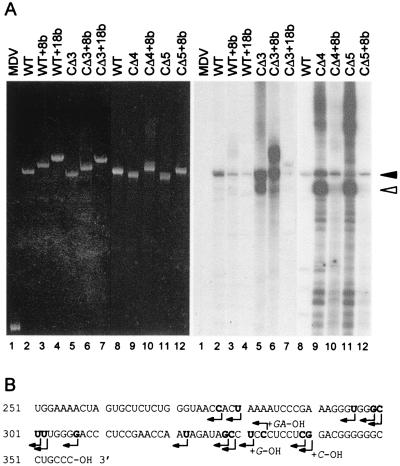
Figure 1.
Transcription of satC templates with or without 3′ end alterations using partially purified TCV RdRp. (A) The templates indicated above each lane were subjected to transcription in vitro using TCV RdRp extracts prepared from infected turnip plants. After denaturing gel electrophoresis, the gel was stained with ethidium bromide to visualize the template RNAs (Left) and subjected to autoradiography to visualize the radiolabeled products (Right). Solid arrow denotes the full-length wt product. The major faster-migrating species is denoted with an open arrow. MDV, 220-base Qβ bacteriophage midivarient RNA; WT, wild-type satC; CΔ3, CΔ4, CΔ5, satC with deletions of three, four, or five bases, respectively. Templates with additional eight or 18 plasmid-derived sequences are denoted by +8b or +18b. (B) Transcription initiation sites (denoted by arrows) of the major products synthesized from templates CΔ3, CΔ4, and CΔ5. Major products (indicated by arrows in A) were gel-purified and cloned before sequence determination (satC-sized product results are shown in Table 3). Nontemplate bases are shown in the (+)-sense orientation to the right of some initiation sites. Initiation sites to the left of adenylates are approximate because the cloning procedure does not allow for the detection of initiation sites at adenylates.