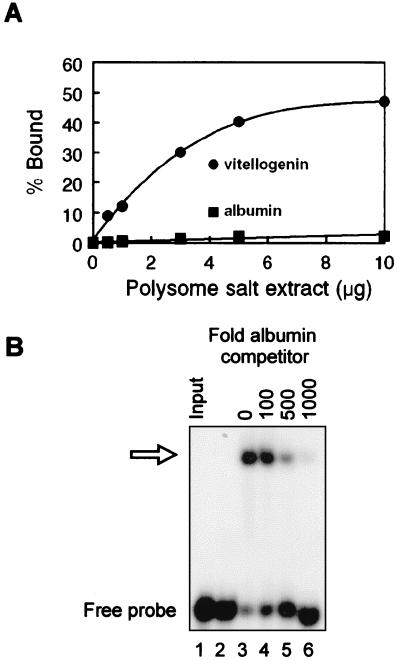
Figure 4.
Vigilin-binding affinity for vitellogenin mRNA 3′-UTR and albumin mRNA. (A) A protein excess-binding curve EMSA was performed by using 32P-labeled albumin mRNA or vitellogenin 3′-UTR and increasing amounts of polysome extract from estrogen-treated frogs. Binding activity was quantified by using the PhosphorImager to compare the relative amount of radiolabeled RNA shifted into slower-migrating complexes versus unbound RNA at the bottom of the gel. (B) The ability of the albumin transcript bearing the PMR-1 cleavage sites to displace vigilin-bound vitellogenin mRNA 3′-UTR was determined by a competition EMSA. Polysome extract (5 μg) from estrogen-treated frogs was incubated with 500 fmol of 32P-labeled vitellogenin mRNA 3′-UTR plus the indicated amounts of unlabeled albumin mRNA competitor. The position of the retarded complex is indicated by the open arrow.