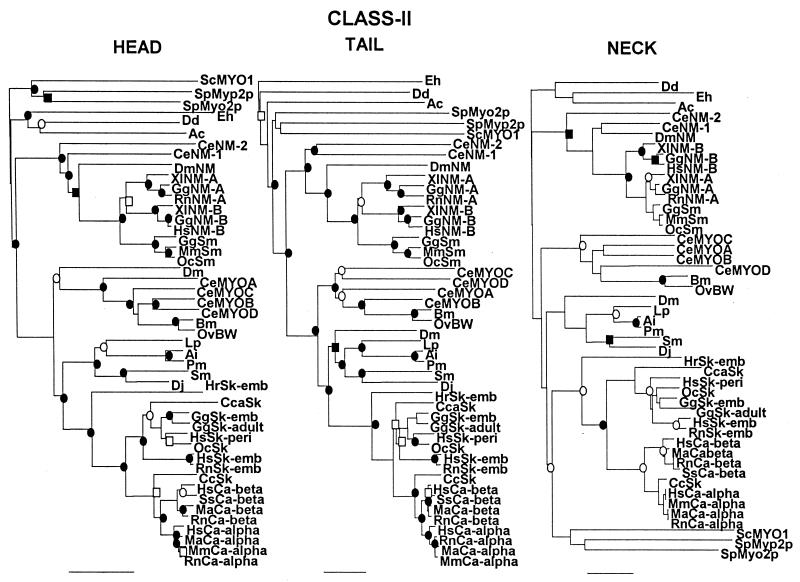
Figure 2.
Comparison of unrooted phylogenetic trees of the head, neck, and tail domains of the heavy chains of class II myosins. These trees are not displayed in radial form (as in Fig. 1) because some of the subgroups are too tightly clustered to be readily visualized in that format. The scale bars of percent divergence are different lengths for the three trees. ●, Nodes found in 100% of bootstrap trials; ○, nodes found in >95% of bootstrap trials; ■, nodes found >90% of bootstrap trials; □, nodes found in >80% of bootstrap trials. The same results were obtained for the neck domains after 10,000 bootstrap trials and when the order of sequence entry was changed. The GenBank accession numbers for each myosin and, in parentheses, the residue numbers used to define the head and neck/tail domains are: Ac, P05659 (1–775, 776–846, 847-1509); Ai, P24733 (1–763, 764–835, 836-1938); Bm, M74000 (1–773, 774–844, 845-1957); CcSk, U53862 (1–761, 762–832, 833-1931); Cca, BAA22069 (1–767, 768–838, 839-1935); CeMYOA, P12844 (1–79, 780–850, 851-1969); CeMYOB, P02566 (1–778, 779–849, 850-1966); CeMYOC, P12845 (1–782, 783–853, 854-1947); CeMYOD, P02567 (1–773, 774–844, 845-1938); CeNM-1, U41990 (1–763, 764–834, 835-1956); CeNM-2, U49263 (1–781, 782–852, 853-2003); Dd, A26655 (1–747, 748–818, 819-2116); Dj, BAA34955 (1–557, 558–628, 629-1743); Dm, A32491 (1–1198, 1199–1259, 1260–2385); DmNM, A36014 (1–815, 816–886, 887-2017); Eh, L03534 (1–780, 781–851, 852-2139); GgNM-A, P14105 (1–764, 765–835, 836-1959); GgNM-B, M93676 (1–802, 803–873, 874-2007); GgSk-emb, P02565 (1–771, 772–842, 843-1940); GgSk-adult, P13538 (1–768, 769–839, 840-1938); GgSm, P10587 (1–777, 778–848, 849-1979); HrSk-emb, D45163 (1–766, 767–837, 838-1927); HsCa-alpha, D00943 (1–768, 769–839, 840-1939); HsCa-beta, P12883 (1–766, 767–837, 838-1935); HsNM-B, M69181 (1–771, 772–842, 843-1976); HsSk-emb, P11055 (1–767, 768–838, 839-1940); HsSk-peri, Z38133 (1–769, 770–840, 841-1937); Lp, AAC24207 (1–765, 766–836, 83701935); MaCa-alpha, L15351 (1–768, 769–839, 840-1939); MaCa-beta, L12104 (1–765, 766–836, 837-1934); MmCa-alpha, M76598 (1–768, 769, 839, 840- 1938); MmSm, D85924 (1–771, 772–842, 843-1938); OcSk, U32574 (1–769, 770–840, 841-1948); OcSm, M77812 (1–771, 772–842, 843-1972); OvBW M74066 (1–773, 774–844, 845-1957); Pm, AAB03660 (1–766, 767–837, 8938–1941); RnCa-alpha, S06005 (1–767, 768–838, 839-1938); RnCa-beta, P02564 (1–766, 767–837, 838-1935); RnNM-A, U41463 (1–764, 765–835, 836-1961); RnSk-emb, P12847 (1–767, 768–838, 835-1940); ScMYO1, NP 011888 (1–779, 780–851, 852-1928); Sm, L01634 (1–752, 753–823, 824-1940); SpMyo2p, AAC04615 (1–755, 756–826, 827-2104); SpMyp2p, U75357 (1–743, 744–814, 815-1526); SsCa-beta, U75316 (1–766, 767- 837, 838-1935); XlNM-A, AAC83556 (1–1-764, 765–835, 836-1964); XlNM-B, A47297 (1–787, 788–858, 859-1992). Abbreviations: As in Fig. 1 and Bm, Btugia malayi (nematode); Cc, Coturnix coturnix (quail); Cca, Cyprinus carpio (carp); Dj, Dugesia japonica (planaria); Hr, Halocynthia roretzi (ascidia); Ma, Mesocritus aureus (hamster); Oc. Oryctolague cuniculus (rabbit); Ov, Onchocerca volvulus (nematode); Pm, Placopecten magellanicus (mollusc); Sm, Schistosoma mansoni; Sp, Schizosaccharomyces pombe; Ss, Sus scrufo (pig); Xl, Xenopus laevis; BW, body wall; Ca, cardiac; emb, embryonic; NM, nonmuscle; peri, perinatal; Sk, skeletal muscle.