Abstract
Based on protein sequence homology searches, we found a conserved open reading frame within the genome of several human pathogenic bacteria showing a resemblance to the mammalian TIR domain. We cloned, expressed and characterized the corresponding gene product from Paracoccus denitrificans using several biophysical techniques. The protein consists of two independently folded domains. As predicted from the amino acid sequence and experimentally confirmed here, the N-terminal domain consists of a α-helical coiled-coil. The NMR data indicates that the C-terminal TIR-like domain folds into a compact protein. Finally, using GST pull-down experiments, we show that the bacteria TIR-like domain binds to the mammalian receptor (TLR4) and adaptor (MyD88) TIR domains. We postulate that prokaryotic pathogens utilize the TIR-like proteins to interfere with the innate immune response of the mammalian host so that the bacterial infection can progress undetected.
Keywords: inflammation, innate immunity, signal transduction, Toll-like receptor pathway, DSC, CD, NMR
Introduction
For protection against pathogens, mammalian hosts have evolved an innate, as opposed to acquired, immune system that can recognize a microbe and mount a response during the first encounter. So far, three protein families of pathogen sensors have been identified: the Toll-like receptors (TLR), the NOD-like receptors and the RIG-like receptors [1] [2]. Mammalian cells use the TLR class, containing a single transmembrane spanning region, to bind to pathogen associated molecular patterns (PAMP), like lipopolysaccharides from Gram negative bacteria. Using the receptor’s intracellular Toll-like/IL-1 receptor/plant Resistance (TIR) domain, the activated receptor recruits a family of cytoplasmic TIR containing proteins known as the adaptors to induce a cascade of signal transduction ultimately leading to the activation of the transcription factor NFκB [3, 4]. As a result, genes involved in organizing an inflammatory response against bacterial infection like TNF-α or interferon-β are switch on.
The TIR domain sequence signature is also present in a family of bacterial proteins [5]. Members include genes from the organisms Staphylococcus aureus, Brucella melitensis, and Salmonella enteriditis. The amino acid sequence shows a conserved domain composition with a predicted coiled-coil region at the N-terminus and a TIR-like domain at the C-terminus. However, the actual role of the bacteria TIR-like proteins (TLP) remains unknown. To date the only reported study for this protein family is from S. enteriditis. It was found that the TlpA protein, encoded in the phage region of the S. enteriditis genome, could indeed suppress the NFκB expression induced by the receptor TLR4 or the adaptor MyD88 in transfected cells [6].
In this paper, we report the identification and biophysical characterization of a bacterial TLP cloned from Paracoccus denitrificans, referred here on as PdTLP (P. denitrificans TIR-like protein). Based on the differential scanning calorimetry (DSC), far UV circular dichroism (CD), and nuclear magnetic resonance (NMR) data, we show that the protein has two domains: an N-terminal α-helical coiled-coil, followed by an independently folded TIR-like domain. Using 35S-methionine labeled proteins in a GST pull-down assay, we observed that the TIR-like domain of PdTLP binds to the human TLR4 and mouse MyD88 TIR domains. We conclude that the aforementioned human pathogens use this class of bacterial TIR-like proteins to inhibit the host’s innate immune signal transduced by the TLR pathway.
Materials and methods
The DNA vectors codifying for the mouse MyD88 and human TLR4 proteins were kindly provided by Dr. Reed (Burnham Institute). The Paracoccus denitrificans strain was purchased from the American Type Culture Collection (catalog number: 13543). The reagents used for DNA cloning and manipulations were obtained from New England Biolabs. All other reagents including proteases were purchased from Sigma-Aldrich.
Gene cloning
The primer design was done according to the NCBI database entry ZP_00632341. The liophylized P. denitrificans strain was rehydrated in 2xTY media (16g/L Tryptone, 10 g/L yeast extract, and 5 g/L NaCl) and grew at 26°C for 48 hours. 1μL of the culture was used as template for PCR. All other PCR were done with pure DNA as template. The restriction digestion and ligation were done according to the manufacturer’s recommended protocol. Escherichia coli XL1Blue strain was used for subcloning and amplification of the ligated vectors. The identity of all constructs was verified by DNA sequencing.
Protein expression
P. denitrificans TIR-like protein (PdTLP) DNA sequence was cloned into a pET15b vector and transformed in the Rosetta DE3 pLysS E. coli strain (Novagen) by CaCl2 method for the purpose of overexpressing the full length protein. The freshly transformed cells were grown in 2xTY (supplemented with 100 μg/mL of ampicillin) until an OD600 of 0.8 was reached at 37°C. 0.2 mM IPTG was added to induce protein expression for 16 hours at 15°C.
Full length PdTLP purification
Cells were harvested by centrifugation at 4°C. Lysis buffer consisting of 20 mM Tris-HCl (tris(hydroxymethyl)aminomethane, titrated with HCl to pH 7.0), 300 mM NaCl and 1 % Triton X-100, was used to resuspend the cell pellet. Cells were lysed by French-press with 3 passes at 1000 PSI and clarified by centrifugation at 43,000 ×g for 1 hour at 4°C. Supernatant was loaded directly into a 5 mL Hi-TRAP Chelating column (GE-Amersham Biosciences), which had been charged with NiCl2 and equilibrated with 50 mL of lysis buffer. Weakly bound proteins were eluted by washing the column with 50 mL of lysis buffer complemented with 30 mM imidazole. The His-tagged protein was eluted by 0.5 M imidazole dissolved in lysis buffer. In order to remove the tag, fractions containing the pure protein were pooled and thrombin (Sigma) added at concentration of 1 unit per mg of protein substrate. The thrombin digestion was incubated with rocking at 4°C for 16 hours. A Sephacryl S200 (GE-Amersham Biosciences) gel filtration column was used to further purify the digested protein. The vector derived residues, Gly-Ser-His, remained attached to the protein after the thrombin cleavage of the N-terminal His-tag.
Limited proteolysis
Full length PdTLP was subjected to mild proteolysis using the proteases trypsin, subtilisin, elastase, thermolysin, and chymotrypsin at a ratio of 1 mg protease : 100 mg protein. The proteolysis was carried out at 4°C for 24 hours. As control a sample with identical concentration of full length PdTLP was prepared and incubated in the same conditions. 5μL of each sample was loaded onto a 4–20% SDS-PAGE (Invitrogen) for analysis. Alternatively, chymotryptic PdTLP fragments were analyzed by mass spectroscopy (Proteomics Facility, Burnham Institute for Medical Research) and N-terminal amino acid sequencing (Protein Chemistry Core Facility, University of Texas).
N-terminal and C-terminal PdTLP fragments purification
The method is similar to that of the full length protein, but after elution from the Ni-column, the full length PdTLP was subjected to controlled proteolysis by chymotrypsin at 4°C for 24 hours. The ratio used for the proteolysis was 1 mg of protease to 1000 mg of purified PdTLP. The chymotrypsin digested sample was buffer-exchanged using a 5 ml Hi-TRAP desalting column from GE-Amersham Biosciences to remove the imidazole. The two fragments were separated by chromatographic methods using Ni-NTA chelating and S200 gel filtration columns.
Differential Scanning Calorimetry
The DSC measurements were carried out with a VP-DSC differential scanning microcalorimeter from MicroCal, LLC (USA). Pure proteins were concentrated to 0.4 mg/mL, and heated at a rate of 1°/min. The protein constructs were buffer-exchanged into 50 mM phosphate and 100 mM NaCl just before data acquisition.
Circular dichroism
The CD experiments were performed using an AVIV 62A spectrometer (Aviv, New Jersey) at a temperature of 15°C. Protein samples were concentrated at 1 mg/mL. A cuvette with a path length of 0.1 mm was used. Wavelengths from 260 to 190 nm were twice scanned with a 0.5 nm interval and an averaging time of 2 seconds. The spectrum shown represents the average signal from both scans after subtraction of the signal obtained with the buffer.
NMR
15N-labelled protein sample was prepared in a similar protocol as described above, except that the expression culture was done in M9 minimal media supplemented with 15N-ammonium chloride (Cambridge Isotope). The purified sample was buffer-exchanged into 10 mM sodium phosphate 300 mM NaCl at pH 7.0. The 1H-15N HSQC experiment was performed at 15°C in a 600 MHz Bruker Avance Spectrometer equipped with Z-gradients and a cryoprobe.
GST-pull down
All glutathione resin beads were equilibrated with 1xPBS buffer before use. The proteins were labeled with 35S-Methionine using the TNT coupled in vitro transcription/translation kit (Promega) following the protocol recommended by the manufacturer. The labeled mixture was pre-cleared with 50μL GSH beads (GE-Amersham Biosciences), before adding to 50μL of GSH beads pre-loaded with GST-hTLR4-TIR or GST-mMyD88-TIR. The mixture was incubated for half hour at room temperature, followed by three washes with 1mL of 1xPBS. Equal volumes of 2x SDS-PAGE running buffer were added and boiled for 5 minutes. The pull-downs were separated on a 4–20% SDS-PAGE (Invitrogen). The gels were dried before exposed to a Kodak Biomax film.
Results
Protein sequence comparisons identify a bacterial family with homology to the mammalian TIR domain
In order to expand the members of the bacterial TIR-like protein (TLP) family, we ran a BLAST search using the sequence of S. enteriditis TLP (TlpA) as template, and found a highly homologous protein from P. denitrificans (NCBI accession number ZP_00632341) which has a 39% sequence similarity over a 255 amino acids range. The supplemental material figure shows the full length sequence alignment between several bacteria TLPs and the TIR domain of three mammalian receptors and an adaptor of the TLR path. At the N-terminal region (top three blocks of the alignment), there is virtually no sequence similarity between the different bacterial TLPs. The consensus secondary structure prediction for this region shows a high α-helical content. However, reliable sequence similarity appears after the F-I/L-S-H motif, which is unique and highly conserved among all bacterial TLPs. This conserved sequence motif aligns with the equivalent F-I/V-S/I/C-Y present in the mammalian TIR proteins as part of the βA strand in the 3D structure (bottom three blocks). The degree of amino acid position conservation or with similar chemical properties of the side chains (~20%), together with the matching of the predicted secondary structure elements in the bacterial sequences with the mammalian ones, makes the likelihood of the presence of the TIR fold in prokaryotes highly reliable.
Limited proteolysis unveils the presence of two domains in PdTLP
Full length PdTLP (299 residues, 32.9 kDa) was expressed as a soluble protein using the Rosetta DE3 pLysS E. coli strain. The protein was purified to high homogeneity using standard Ni affinity and gel filtration chromatography, with a yield of about 20 mg/L of culture. The molecular weight of the purified protein was confirmed by mass spectrometry. Given that we are working with a predicted multidomain protein, limited proteolysis is a powerful technique to determine the domain boundaries and the stability of protein fragments in solution. Five proteases with different chemical specificities were selected for limited digestion of the full length PdTLP. As shown in Figure 1A, we found that PdTLP can be digested to at least two fragments by all tested proteases. Chymotrypsin produced the simplest proteolytic digestion pattern consisting of a single fragment of apparent molecular weight of 16 kDa as judged by SDS-PAGE. N-terminal amino acid sequencing of this fragment provided the sequence: Ser-Ala-Met-Lys-Pro. Together with the mass spectrometry data obtained from the same fragment permitted its unequivocal identification as corresponding to the C-terminal region, from Ser146 to Asp299. The N-terminal region was not detected in chymotrypsin treated samples indicating that is rapidly digested to multiple peptides and suggesting it adopts a less stable fold. In order to purify both fragments, we optimised the digestion conditions and scaled up the production. After incubation of the purified full length protein with the protease chymotrypsin, the His-tagged N-terminal coiled-coil domain was bound to the Ni-column, while the TIR-like domain was collected in the flow through achieving the separation of both polypeptides (Figure 1B). Based on the elution volume of the gel filtration chromatograph, the full length protein behaves as a dimer, while the TIR-like domain elutes at the position expected for a monomer (Figure 1C). The coiled-coil domain exists as a mixture of predominantly monomers as well as in higher oligomeric states.
Figure 1.
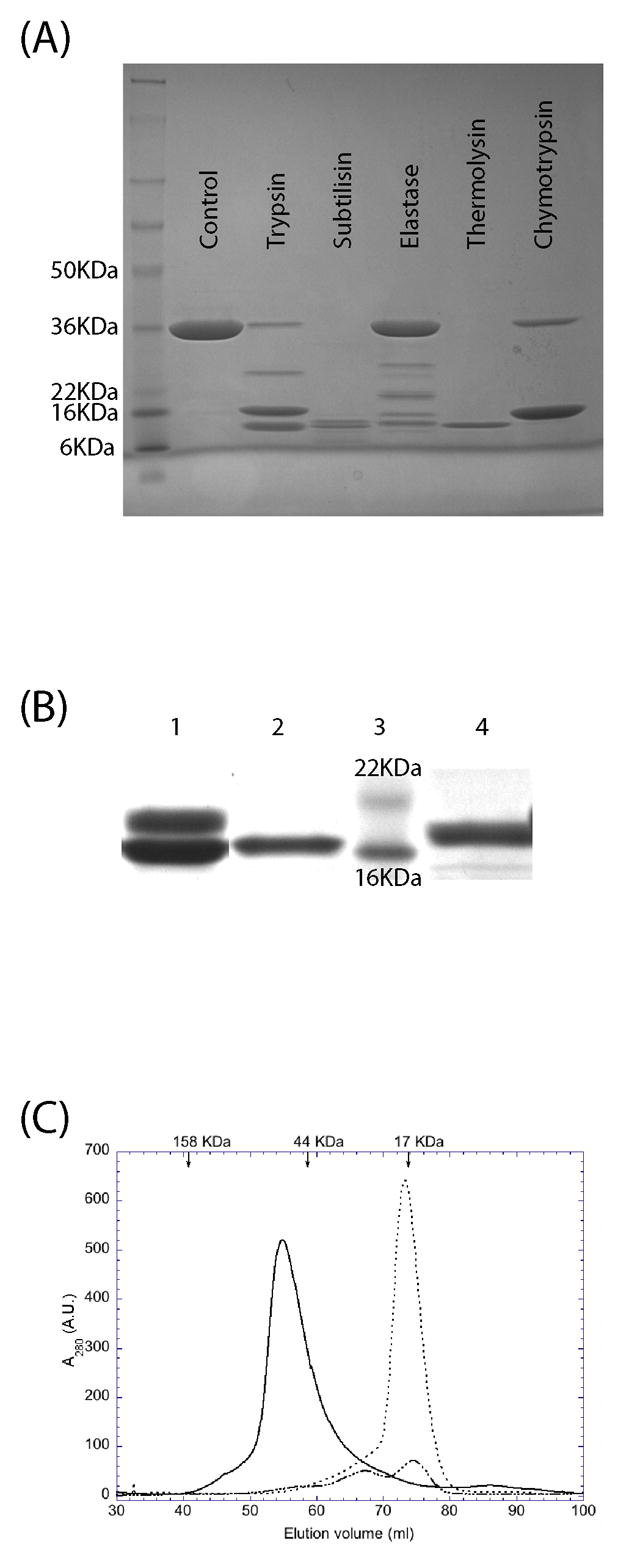
Purification and limited proteolysis analysis of PdTLP. (A) SDS-PAGE of the limited proteolysis experiments stained with coomassie blue. (B) SDS-PAGE of the scaled-up preparation of coiled-coil and TIR-like domains of PdTLP. Lane 1: Chymotrypsin treated PdTLP giving two bands corresponding to the two domains; Lane 2: Ni-column flow through; Lane 3: molecular weight markers; Lane 4: imidazole eluted fraction from the Ni-column. (C) Gel filtration chromatography profile of the PdTLP full length (___), coiled-coil (-.-), and TIR-like (…) domains. Arrows at the top indicate the eluted position of the molecular weight markers.
PdTLP is composed of two independently folded domains
DSC provides information regarding the thermal stability as well as the potential molecular interactions between proteins or domains. The DSC profiles of the full length, N- and C-terminal domains of PdTLP are shown in Figure 2A. Table 1 contains the measured thermodynamic parameters for each polypeptide. The full length construct displayed a two-peak heat release curve, demonstrating that there were two thermal unfolding events. The first peak is an almost perfect fit with the melting curve of the N-terminal domain suggesting that, the first thermal event observed for the full length PdTLP reflects mainly the unfolding of the N-terminal domain. This matching also indicates that the two domains interact weakly with each other in the context of the full length protein. The second event in the unfolding of the full length protein has a melting temperature almost five degrees lower than that of the melting temperature of the C-terminal domain. We reasoned that this might be due to the destabilizing effect of the attached, and at that temperature unfolded, N-terminal domain. Without the N-terminal domain, the C-terminal domain is more stable. Two peaks were observed from the thermal unfolding of the C-terminal domain alone, even after repeating the experiment several times with different samples coming from separated purifications and digestions. Even though the majority of the protein has a melting temperature of 63.5 °C, we speculate that the minor peak at around 47 °C comes from the unfolding of an alternate and less stable conformation populated by a minority of molecules.
Figure 2.
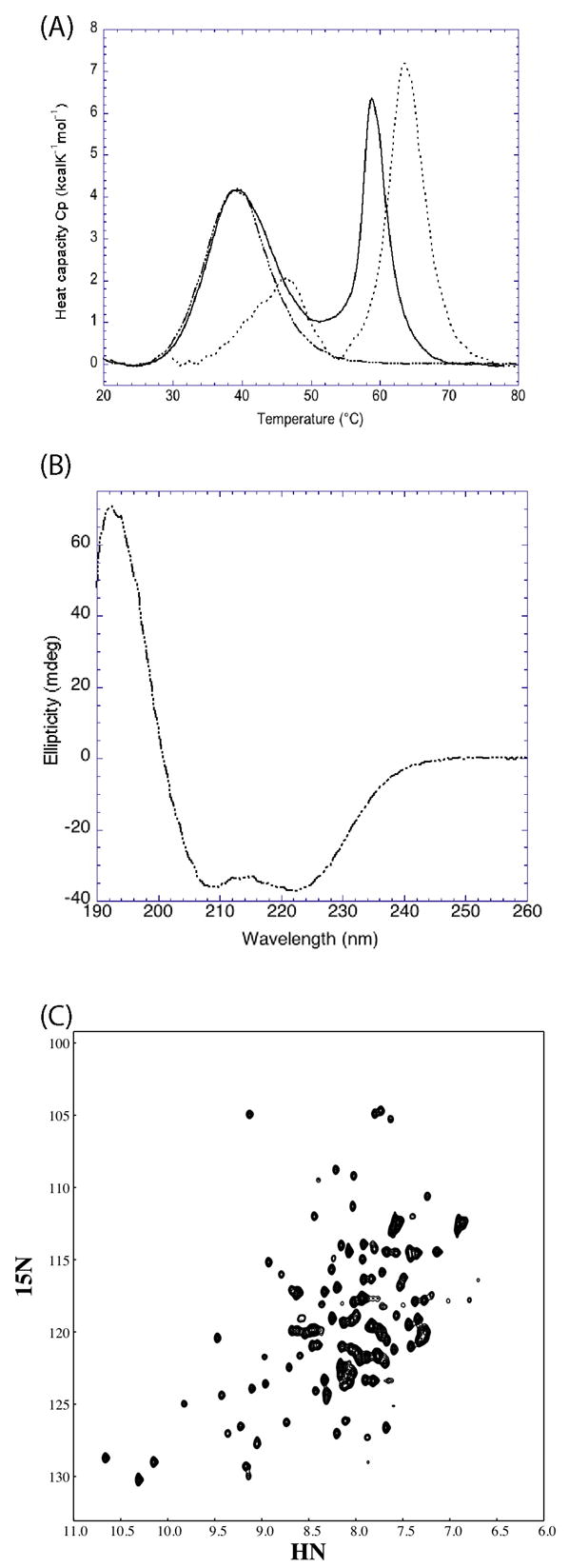
Biophysical characterization of PdTLP. (A) Differential scanning calorimetry profiles of the full length (___), coiled-coil (-.-) and TIR-like (…) domains of PdTLP. (B) Far UV circular dichroism spectrum of the coiled-coil domain of PdTLP. (C) 1H-15N HSQC spectrum of PdTLP TIR-like domain. The units in the x- and y- axis are parts per million (ppm).
Table 1.
Thermodynamic parameters obtained from the DSC experiments of PdTLP.
| Tm1 (°C) | Tm2 (°C) | ΔH (kcal/mol) | |
|---|---|---|---|
| Full length | 38.7 | 58.8 | 88.1 |
| Coiled-coil domain | 39.3 | - | 65.1 |
| TIR-like domain | - | 63.5 | 71.3 |
PdTLP N-terminal region folds into a α-helical coiled-coil domain
Given that the secondary structure for the N-terminal region is predicted to be mainly α-helical, we used circular dichroism to test it. The far UV circular dichroism spectrum for the N-terminal fragment of PdTLP is shown in figure 2B. As expected, we found that the N-terminal domain has a high α-helical content with the typical double minimal signal with negative ellipticity values at 222 and 208 nm followed by a positive maximum at 192 nm. This result is compatible with the region adopting a coiled-coil conformation.
PdTLP TIR-like domain folds independently of the rest of the protein
Figure 2C shows the two-dimensional 1H-15N HSQC NMR spectrum of the C-terminal domain of PdTLP. The spectrum contains sharp and well dispersed resonances as expected for a compact folded domain. The number of cross peak is almost equivalent to the number of amino acids present, and the full assignment of the spectrum is underway.
Bacteria TIR-like domains interact with mammalian TIR domains
We used GST pull-down experiments to determine if there is any interaction between the TIR-like domain of PdTLP and the human TLR4 and mouse MyD88 TIR domains. We found that the 35S-Methionine labeled PdTLP TIR-like and mMyD88 TIR fragments were pull-down by both GST-mMyD88-TIR and GST-hTLR4-TIR (Figure 3). Even though the intensity of the bands is rather low, suggesting a relatively weak binding, this experiment clearly shows that the PdTLP TIR-like domain specifically binds to mammalian receptor and adaptor TIR containing proteins.
Figure 3.
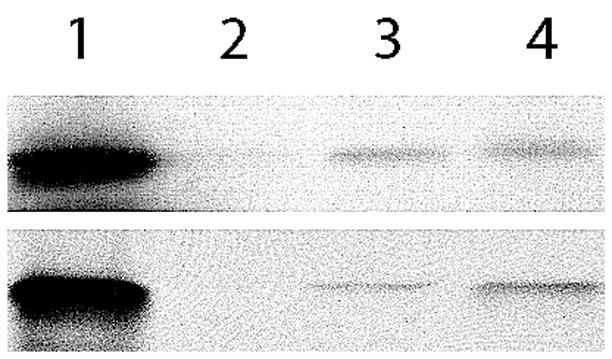
GST pull-down experiments. Top gel shows the 35S-labelled PdTLP TIR-like (lane 1) followed by the pull-down of both GST-mMyD88-TIR (3) and GST-hTLR4-TIR (4), but not GST alone (2). Bottom gel is the pull-down signal coming from the positive control mMyD88 TIR.
Discussion
Protein sequence comparisons of mammalian TIR domains with a prokaryote genome database permitted the identification of a family of bacterial proteins with significant homology to the receptors and adaptors of the Toll-like receptor signaling pathway. Furthermore, the gene location of the TIR-like proteins (TLP) in S. enteriditis, S. aureus or B. melitensis is inside pathogenicity islands of phage origin containing genes coding for DNA integrases and type IV secretion pilin protein precursors [7]. Such position inside the genome suggests that TLPs could function as virulent factors, as it is the case for other infectious proteins [8]. Our work has focused on the TLP from P. denitrificans (PdTLP). The limited proteolysis and DSC experiments performed with the entire polypeptide showed that the PdTLP is made up of two independently folded domains separated by a flexible linker. Corroborating the predicted secondary structure, the far UV CD spectrum of the N-terminal domain has the characteristic signature of a α-helical coiled-coil domain. The 1H-15N HSQC NMR spectrum of the C-terminal TIR-like domain displays the typical signal dispersion expected for a well folded protein. Investigating TLPs signal transduction properties [9], GST-pull down experiments demonstrate a physical interaction between prokaryote TIR-like domains and their mammalian counterparts, TIR domains from both receptors and adaptors of the TLR path.
Taken together, the results described here permit us to advance the following model of how the bacterial TIR-like proteins may inhibit signals coming from the mammalian TIR adaptor proteins (Figure 4). We showed, by gel filtration chromatography that the PdTLP behaves as a dimer. A plausible implication is that the dimer conformation of the full length protein mimics the activated dimer form of the mammalian Toll-like receptor such that it can bind and sequester the main adaptor, the TIR containing protein MyD88 [10]. It is also known that MyD88 forms dimers in mammalian cells [11]. The dimeric PdTLP could imitate such a state too. Given the ability to directly bind TLR4 and MyD88 as observed in the pull-down assays, the PdTLP may promote unproductive complexes of TIR proteins in infected cells, thereby blocking the TLR signal transmission and ultimately preventing the activation of NFκB. In this way, even though cell death is occurring [6], the infected cell will not send out interleukins that might alert the host organism to mount an innate immune response against the pathogen.
Figure 4.
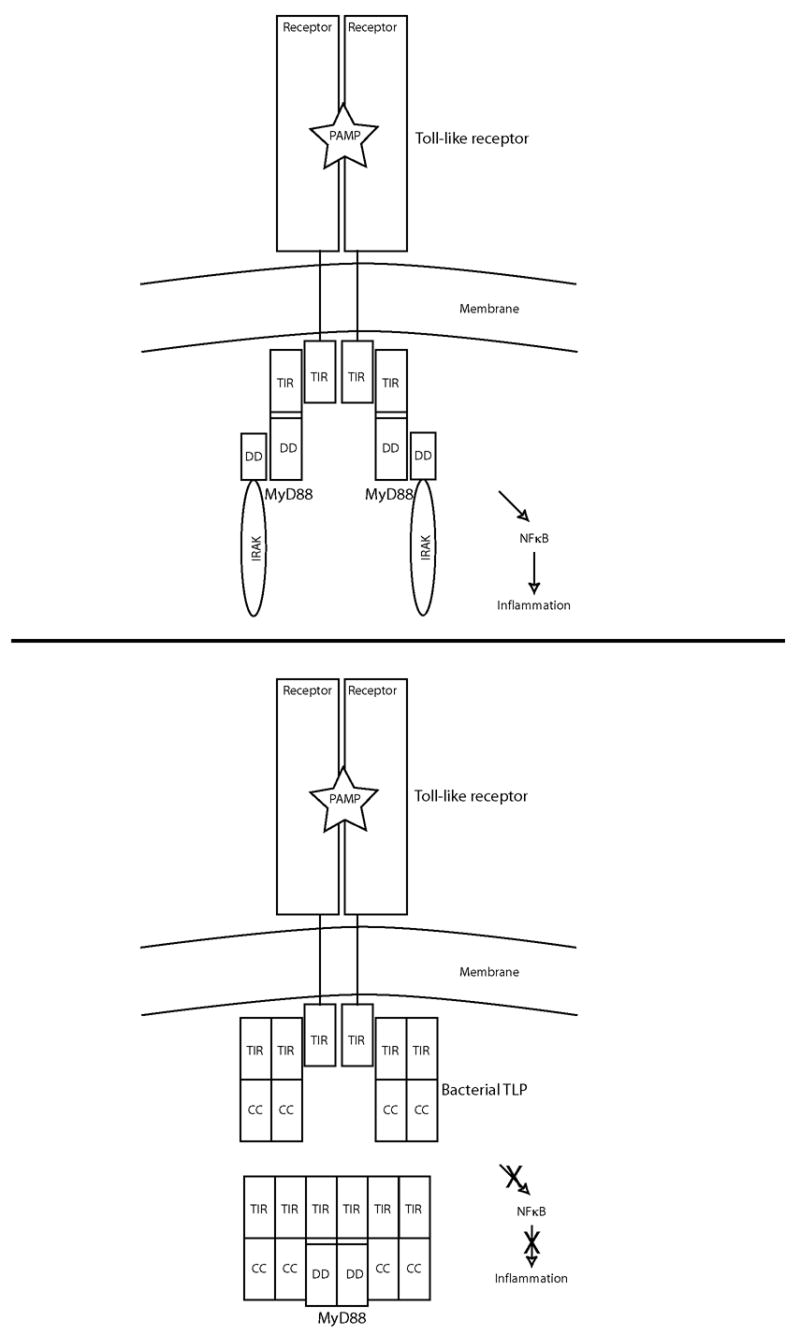
A model of how bacterial pathogens may use the TIR-like proteins to inhibit NFκB signaling in infected cells. Top panel: a simplified depiction of the Toll-like receptor signaling pathway with a productive receptor-adaptor TIR-TIR interaction. Bottom panel: the presence of bacterial TLPs, prevents the inflammatory signal to proceed by the formation of a bacterial-host TIR-TIR interaction. Legend: PAMP, pathogen-associated molecular patterns; TIR, Toll/IL-1 receptor/plant Resistance domain; DD, death domain; CC, coiled-coil.
Several cases of structural and functional mimicry between virulent factors and host signaling proteins have been reported [12] [13]. In our case, given the significant degree of homology is expected that both the bacterial TIR-like and the mammalian TIR domains share very similar 3D fold and participate in the same protein-protein interactions. Therefore, the bacterial TIR-like proteins may be competing with the TIR containing adaptors for complex formation with the Toll-like receptors. In this manner the host innate immune system could be silenced, allowing the bacteria to proceed with the infection within the host cell undetected. This would be a novel mechanism by which the pathogen could attenuate and manipulate its host immune response. Ultimately, it could provide a target for drug design in order to treat bacterial infection [14].
Supplementary Material
Sequence alignment of selected TIR containing bacteria and mammalian proteins obtained with the software Clustal-X. The numbering follows the PdTLP sequence, and the position of the secondary structure elements is either predicted for the coiled-coil region (labeled as α-helix) or based on the structure of human TLR1 TIR domain for the TIR-like region. PdTLP, BmTLP, TlpA, SaTLP and EcTLP identify the sequences from Paracoccus denitrificans (ZP_00632341), Brucella melitensis (NP_540591), Salmonella enteriditis (UIUC_592), Staphylococcus aureus (YP_042171) and Escherichia coli (NP_754290), respectively. The asterisks, colons and period signs mark conserved, partially conserved or chemical property conserved positions.
Acknowledgments
We would like to thank Dr Andrey Bobkov for collecting the DSC data and Prof. Robert C. Liddington and his research group for helpful suggestions throughout the project. This work was funded by grants R21AI065602 and P01AI055789 to JP.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Creagh EM, O’Neill LA. TLRs, NLRs and RLRs: a trinity of pathogen sensors that co-operate in innate immunity. Trends Immunol. 2006;27:352–357. doi: 10.1016/j.it.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 2.Werts C, Girardin SE, Philpott DJ. TIR, CARD and PYRIN: three domains for an antimicrobial triad. Cell Death Differ. 2006;13:798–815. doi: 10.1038/sj.cdd.4401890. [DOI] [PubMed] [Google Scholar]
- 3.Doyle SL, O’Neill LA. Toll-like receptors: from the discovery of NFkappaB to new insights into transcriptional regulations in innate immunity. Biochem Pharmacol. 2006;72:1102–1113. doi: 10.1016/j.bcp.2006.07.010. [DOI] [PubMed] [Google Scholar]
- 4.Lasker MV, Nair SK. Intracellular TLR signaling: a structural perspective on human disease. J Immunol. 2006;177:11–16. doi: 10.4049/jimmunol.177.1.11. [DOI] [PubMed] [Google Scholar]
- 5.Koonin EV, Aravind L. Origin and evolution of eukaryotic apoptosis: the bacterial connection. Cell Death Differ. 2002;9:394–404. doi: 10.1038/sj.cdd.4400991. [DOI] [PubMed] [Google Scholar]
- 6.Newman RM, Salunkhe P, Godzik A, Reed JC. Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect Immun. 2006;74:594–601. doi: 10.1128/IAI.74.1.594-601.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.McNeil LK, Reich C, Aziz RK, Bartels D, Cohoon M, Disz T, Edwards RA, Gerdes S, Hwang K, Kubal M, Margaryan GR, Meyer F, Mihalo W, Olsen GJ, Olson R, Osterman A, Paarmann D, Paczian T, Parrello B, Pusch GD, Rodionov DA, Shi X, Vassieva O, Vonstein V, Zagnitko O, Xia F, Zinner J, Overbeek R, Stevens R. The National Microbial Pathogen Database Resource (NMPDR): a genomics platform based on subsystem annotation. Nucleic Acids Res. 2007;35:D347–353. doi: 10.1093/nar/gkl947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gal-Mor O, Finlay BB. Pathogenicity islands: a molecular toolbox for bacterial virulence. Cell Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 9.Brown V, Brown RA, Ozinsky A, Hesselberth JR, Fields S. Binding specificity of Toll-like receptor cytoplasmic domains. Eur J Immunol. 2006;36:742–753. doi: 10.1002/eji.200535158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gay NJ, Gangloff M, Weber AN. Toll-like receptors as molecular switches. Nat Rev Immunol. 2006;6:693–698. doi: 10.1038/nri1916. [DOI] [PubMed] [Google Scholar]
- 11.Burns K, Martinon F, Esslinger C, Pahl H, Schneider P, Bodmer JL, Di Marco F, French L, Tschopp J. MyD88, an adapter protein involved in interleukin-1 signaling. J Biol Chem. 1998;273:12203–12209. doi: 10.1074/jbc.273.20.12203. [DOI] [PubMed] [Google Scholar]
- 12.Prehna G, Ivanov MI, Bliska JB, Stebbins CE. Yersinia virulence depends on mimicry of host rho-family nucleotide dissociation inhibitors. Cell. 2006;126:869–880. doi: 10.1016/j.cell.2006.06.056. [DOI] [PubMed] [Google Scholar]
- 13.Stebbins CE, Galan JE. Structural mimicry in bacterial virulence. Nature. 2001;412:701–705. doi: 10.1038/35089000. [DOI] [PubMed] [Google Scholar]
- 14.O’Neill LA. Targeting signal transduction as a strategy to treat inflammatory diseases. Nat Rev Drug Discov. 2006;5:549–563. doi: 10.1038/nrd2070. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sequence alignment of selected TIR containing bacteria and mammalian proteins obtained with the software Clustal-X. The numbering follows the PdTLP sequence, and the position of the secondary structure elements is either predicted for the coiled-coil region (labeled as α-helix) or based on the structure of human TLR1 TIR domain for the TIR-like region. PdTLP, BmTLP, TlpA, SaTLP and EcTLP identify the sequences from Paracoccus denitrificans (ZP_00632341), Brucella melitensis (NP_540591), Salmonella enteriditis (UIUC_592), Staphylococcus aureus (YP_042171) and Escherichia coli (NP_754290), respectively. The asterisks, colons and period signs mark conserved, partially conserved or chemical property conserved positions.


