Abstract
Aims
The aim of this study was to develop and validate a population pharmacokinetic model of ritonavir, used as an antiviral agent or as a booster, in a large patient population and to identify factors influencing its pharmacokinetics.
Methods
Ambulatory HIV-1-infected patients from the outpatient clinic of the Slotervaart Hospital, Amsterdam, the Netherlands, who were being treated with a ritonavir-containing regimen were included. During regular visits, blood samples were collected for the determination of ritonavir plasma concentrations and several clinical chemistry parameters. Furthermore, complete pharmacokinetic curves were available in some patients. Single and multiple compartment models with zero-order and first-order absorption, with and without absorption lag-time, with linear and nonlinear elimination were tested, using nonlinear mixed effect modelling (NONMEM). Pharmacokinetic parameters and interindividual, interoccasion and residual variability were estimated. In addition, the influence of several factors (e.g. patient characteristics, comedication) on the pharmacokinetics of ritonavir was explored.
Results
From 186 patients 505 ritonavir plasma concentrations at a single time-point and 55 full pharmacokinetic profiles were available, resulting in a database of 1228 plasma ritonavir concentrations. In total 62% of the patients used ritonavir as a booster of their protease inhibitor containing antiretroviral regimen. First order absorption in combination with one-compartment disposition best described the pharmacokinetics of ritonavir. Clearance, volume of distribution and absorption rate constant were 10.5 l h−1 (95% prediction interval (95% PI) 9.38–11.7), 96.6 l (95% PI 67.2–121) and 0.871 h−1 (95% PI 0.429–1.47), respectively, with 38.3%, 80.0% and 169% interindividual variability, respectively. The interoccasion variability in the apparent bioavailability was 59.1%. The concomitant use of lopinavir resulted in a 2.7-fold increase in the clearance of ritonavir (P value < 0.001). No patients characteristics influenced the pharmacokinetics of ritonavir.
Conclusions
The pharmacokinetic parameters of ritonavir were adequately described by our population pharmacokinetic model. Concomitant use of the protease inhibitor lopinavir strongly influenced the pharmacokinetics of ritonavir. The model has been validated and can be used for further investigation of the interaction between ritonavir and other protease inhibitors.
Keywords: pharmacokinetics, ritonavir
Introduction
Ritonavir is a potent human immunodeficiency virus (HIV) protease inhibitor and shows effective antiretroviral activity. However, the use of ritonavir in therapeutic doses is limited by neurological and gastrointestinal toxicity [1]. Both antiviral activity and side-effects have been correlated with plasma ritonavir concentrations [1–3].
Initially, ritonavir was used for its antiviral effect (twice daily 600 mg) in combination with other antiretroviral drugs [3]. However, it was soon recognized that ritonavir in a low dose improves the pharmacokinetic profile of the different co-administered protease inhibitors by raising their concentrations in plasma, increasing their elimination half-lives and reducing the influence of food on their gastrointestinal absorption. Ritonavir has revolutionized antiretroviral therapy and a large increase in the use of low-dose ritonavir in combination with a variety of protease inhibitors has occurred since its introduction [4].
Potent inhibition of cytochrome P450 (CYP) 3A4-mediated metabolism in the gut wall and liver by ritonavir results in the desirable drug–drug interactions with other protease inhibitors [4–9]. Ritonavir also inhibits CYP2D6-mediated metabolism, and to a lesser extent CYP2C9, CYP2C19 and CYP1A2 [10, 11]. In addition, ritonavir may induce the activity of CYP1A2 and glucuronosyl transferase, and possibly CYP2C9 and CYP2C19 [12, 13]. Ritonavir is also an inhibitor of the drug transporter P-glycoprotein (Pgp) and/or the multidrug resistance-associated protein (MRP1). This might result in increased absorption, decreased elimination and improved retention into viral sanctuary sites of other protease inhibitors [14, 15].
The maintenance of high plasma concentrations of protease inhibitors is associated with a more potent and durable suppression of viral replication and with a delay in the development of resistance [16]. Thus, the large increase in the plasma concentrations of other protease inhibitors when co-administered with ritonavir forms the basis of rational dual protease inhibitor regimens.
Few data on the pharmacokinetics of ritonavir are available [13, 17–21]. Furthermore, most studies [17–19] have been executed with HIV-negative subjects and/or before steady-state pharmacokinetic conditions were reached.
Because of several potential drug–drug interactions with ritonavir and variability in the expression of CYP enzymes, Pgp and MRP1, the pharmacokinetics of ritonavir may be complex. Knowledge about their variability may be of help in understanding differences in antiretroviral activity and side-effects, and in gaining more insight into the interaction between ritonavir and other protease inhibitors.
Therefore, the aim of this study was to characterize the population pharmacokinetics of ritonavir, both used as a therapeutic antiretroviral drug or as a booster, in a large and representative patient population, in which various dosages were being used. Furthermore, patient characteristics and other factors influencing the pharmacokinetics of ritonavir were investigated.
Methods
Patients
Subjects were ambulatory HIV-1-infected patients from the outpatient clinic of the Slotervaart Hospital, Amsterdam, the Netherlands. Data were collected during regular outpatient visits, between January 1999 and June 2003. Each visit of the patient to the clinic was considered as an occasion. All patients were using ritonavir as part of their antiretroviral regimen and had at least one plasma ritonavir concentration available for analysis. Patients received ritonavir as a booster or as a therapeutic drug. When patients had a ritonavir plasma concentration below 0.01 mg l−1, they were excluded from pharmacokinetic analysis because of questionable adherence to therapy [22]. In addition to the random samples, full pharmacokinetic profiles (12–15 time points) were available from 55 patients, which were collected as part of several studies performed in our hospital [5, 23–26]. The rationale for pooled analysis is to increase the power of the study. Combining plasma concentration time points from several studies with random samples resulted in a large robust data set. When data from studies are used separately to develop pharmacokinetic models, parameters may be estimated with less precision. Furthermore, a small data set would contain less variability in patient characteristics and factors contributing to the interindividual variability may be hard to detect. Study protocols were approved by the institutional committee on medical ethics and informed consent was obtained from all patients. Single blood samples were obtained during regular follow-up of HIV-infected patients in our hospital according to local treatment guidelines and ethical approval.
Sampling and bioanalysis
At each visit to the clinic, a blood sample was obtained for the determination of plasma ritonavir concentration. Within the therapeutic drug monitoring program in our hospital, a strict protocol is utilized in which plasma concentrations of antiretroviral drugs are routinely and frequently monitored. As a consequence, patients are conversant with the principle of recording time of ingestion of the last dose. Additionally, sampling times are recorded electronically at the Department of Clinical Chemistry. From this information, time after ingestion was estimated. All samples were collected at steady state, at least 2 weeks after initiation of a ritonavir containing regimen.
Plasma concentrations of ritonavir were determined using an isocratic reversed-phase ion-pair, high-performance liquid chromatographic (HPLC) assay with ultraviolet detection (UV) at 239 nm [27]. This method was validated over the range 0.05–25 mg l−1 using 600 µl of plasma. The assay was precise and accurate with between-day and within-day variation of quality control samples of ritonavir in plasma ranging from 0.7 to 7.6%. The mean accuracy was 104.0%.
Covariates
To identify possible relationships between the pharmacokinetics of ritonavir and patient characteristics, data on the following variables were collected at baseline: gender, race, alanine aminotransferase (ALAT, U l−1), aspartate aminotransferase (ASAT, U l−1), alkaline phosphatase (AP, U l−1), gamma-glutamyltransferase (GGT, U l−1), total bilirubin (TBR, µmol l−1), CD4 cells (/mm3), CD8 cells (/mm3) and HIV viral load (number of copies ml−1). Patients were considered to have a chronic hepatitis B infection when hepatitis surface antigen (HbsAg) could be detected at baseline. When antihepatitis C antibodies (anti-HCV) were present at baseline, patients were considered to have a chronic hepatitis C infection. In addition, during treatment with ritonavir data on the following covariates were collected: age (years), body weight (kg), serum creatinine (µmol l−1). The effects of concomitant use of lopinavir, saquinavir and indinavir were also investigated. CD4 cells, CD8 cells, viral load, age, weight and serum creatinine were examined as continuous variables. Gender, race, hepatitis B and hepatitis C infection were examined as dichotomous variables. The values of ALAT, ASAT, AP, GGT and TBR were transformed to dichotomous variables by using 1.5 times the upper limit of normal for AP, GGT and TBR, and 2 times the upper limit of normal for ASAT and ALAT as cut-off values. Not all variables were available from all patients.
Population pharmacokinetic analyses
The nonlinear mixed effect modelling software program NONMEM (version V, level 1.1, GloboMax LLC, Hanover MD, USA), using a Fortran compiler (Compaq Visual Fortran Version 6.5, Compaq Computer Corporation, Houston, TX, USA), was used to perform the analyses. The first-order conditional estimation (FOCE) procedure was used throughout. The INTERACTION option was used to account for interaction between the interindividual, intraindividual and residual error. The adequacy of the developed structural models was evaluated using both statistical and graphical methods. The minimal value of the objective function (OFV) provided by NONMEM was used for the comparisons of the nested models. Discrimination between these hierarchical models was based on the OFV using the log-likelihood ratio test [28]. A P value of 0.05, representing a decrease in OFV of 3.84 was considered statistically significant (chi-square distribution, degrees of freedom (d.f) = 1).
Standard errors for all parameters were approximated using the COVARIANCE option of NONMEM. Individual Bayesian pharmacokinetic estimates of the pharmacokinetic parameters were obtained using the POSTHOC option [28].
Basic pharmacokinetic model
Zero-order and first-order absorption models with and without absorption lag-time were tested. To describe the distribution kinetics of ritonavir, single and multiple compartment models with linear and nonlinear elimination were investigated.
Population pharmacokinetic parameters such as clearance, volume of distribution and absorption rate constant were estimated. Interindividual (IIV) and interoccasion variability (IOV) in the pharmacokinetic parameters and in the apparent bioavailability (F) were estimated from an exponential error model, according to Karlsson & Sheiner [29]. For instance, variability in clearance was determined from the equation:
in which CL/Fij represents the clearance of the ith individual on the jth occasion, θ1 is the typical value of clearance, ηi is the interindividual random effect with a mean of 0 and variance ω2, and κj is the interoccasion random effect with a mean of 0 and variance π2. Residual variability was modelled with a combined additive and proportional error model. Subpopulations were estimated using the $MIX function in the control stream.
Covariate pharmacokinetic model
To identify factors influencing the pharmacokinetics of ritonavir, covariates were introduced separately into the basic model. Covariates were also incorporated into the model to determine the influence of missing data and to avoid bias. For instance, the influence of a dichotomous covariate X on clearance with missing data of X for some individuals was modelled using the equation:
in which TVCL is the typical value of clearance in the population, MIS is equal to 1 for records with missing data and 0 for all other records, θ1 is the typical value of an individual with X = 0 (no missing data), θ2 is the relative difference in clearance for individuals with X = 1 (no missing data) and θ3 is the relative difference in clearance for individuals with missing data.
A covariate was considered statistically significant when the inclusion was associated with a decrease in OFV associated with a P value of ≤0.05 (log-likelihood ratio test). Clinical relevance was assumed when the typical value of the pharmacokinetic parameter of interest changed at least 10% in the range of the covariate observed in the population in order to prevent the detection of an irrelevant albeit significant relationship.
All significant and relevant covariates were included in an intermediate model. Finally, a stepwise backward elimination procedure was carried out. A covariate was retained in the model when the influence of this parameter was statistically significant (P < 0.05) and clinically relevant (10% change in pharmacokinetic parameter).
Statistical refinement
The validity of the interindividual and interoccasion variability model was assessed by evaluating correlations between individual random effects (η) and interoccasion random effects (κ) for all of the pharmacokinetic parameters [30]. When a substantial correlation was present, covariance between these parameters was included in the model.
Model validation
The bootstrap resampling technique was applied as an internal validation. Bootstrap replicates were generated by sampling randomly approximately 65% from the original data set with replacement [31]. The final model was fitted to the replicate data sets using the bootstrap option in the software package Wings for NONMEM (by N. Holford, version 222, May 2001, Auckland, New Zealand) and parameter estimates for each of the replicate data sets were obtained. The precision of the model was evaluated by visual inspection of the distribution of the model parameters. Furthermore, the median parameter values and 95% prediction intervals of the bootstrap replicates were compared with the estimates of the original data set.
Results
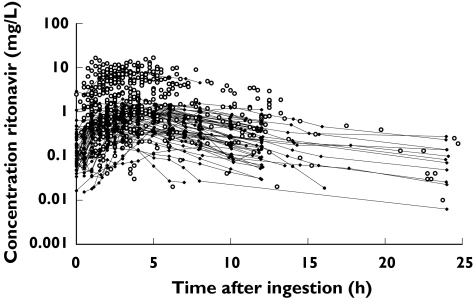
From 186 outpatients, 55 full pharmacokinetic profiles and 505 plasma concentrations at a single time point were available, resulting in a database of 1228 plasma ritonavir concentrations. A total of 115 patients received 100 mg ritonavir once a day or 100 mg, 133 mg or 200 mg ritonavir twice a day as a booster. A total of 71 patients received ritonavir as an antiviral drug in a dosage of 300 mg, 400 mg, 500 mg, 600 mg or 750 mg twice daily. When the full profiles were not taken into account, average 3–4 samples (over a follow-up of 7–12 months) per patient (ranging from 1 to 15, i.e. follow-up up to 28 months) were available. Figure 1 shows all the concentration-time data for ritonavir. The patient population was predominantly male and Caucasian. Demographics and other patient characteristics were not available from 0–37% of the patients (depending on the covariable). In most cases covariates were missing nonrandomly. Thus when one covariate was missing, there was a high probability all covariates were missing for that patient. This limited the opportunities to use joint-modelling or multiple imputations as techniques for dealing with missing data [32]. The characteristics of the patients studied are presented in Table 1.
Figure 1.
Concentration-time data for ritonavir. (○) plasma concentrations at a single time point, dots connected with hairlines (•) full pharmacokinetic profiles
Table 1.
Characteristics of the 186 patients studied
| Parameter | Median | IQR | Missing (n, %) | |
|---|---|---|---|---|
| Regimen | ||||
| Ritonavir (therapeutic (n, %)) | 71 (38.2) | |||
| Ritonavir (booster (n, %)) | 115 (61.8) | |||
| Indinavir/Ritonavir (therapeutic (n, %)) | 9 (4.8) | |||
| Indinavir/Ritonavir (booster (n, %)) | 40 (21.5) | |||
| Saquinavir/Ritonavir (therapeutic (n, %)) | 39 (21.0) | |||
| Saquinavir/Ritonavir (booster (n, %)) | 39 (21.0) | |||
| Lopinavir/Ritonavir (booster (n, %)) | 36 (19.4) | |||
| Age (years) | 39.4 | 35.0–46.0 | 0 (0) | |
| Gender M/F (n, %) | 146/23 (78.5/12.4) | 17 (9.1) | ||
| Weight (kg) | 71.5 | 63.0–79.8 | 32 (17.2) | |
| Race | 29 (15.6) | |||
| Caucasian (n, %) | 120 (64.5) | |||
| Black (n, %) | 19 (10.2) | |||
| Asian (n, %) | 8 (4.3) | |||
| Latino (n, %) | 10 (5.4) | |||
| Clinical chemistry | ||||
| Baseline ASAT (U l−1) | 32 | 27–46 | 44 (23.7) | |
| Baseline ALAT (U l−1) | 40 | 28–52 | 44 (23.7) | |
| Baseline GGT (U l−1) | 30 | 20–61 | 69 (37.1) | |
| Baseline AP (U l−1) | 76 | 62–93 | 45 (24.2) | |
| Baseline TBR (µmol l−1) | 11 | 9–15 | 55 (29.6) | |
| Clinical immunology at baseline | ||||
| CD4 cell count (106 l−1) | 240 | 110–380 | 49 (26.3) | |
| CD8 cell count (106 l−1) | 960 | 580–1360 | 49 (26.3) | |
| Molecular biology at baseline | ||||
| Plasma log10 HIV-1 RNA (copies ml−1) | 4.86 | 3.48–5.47 | 40 (21.5) | |
| HBV/no HBV (n, %) | 7/138 (3.8/74.2) | 41 (22.0) | ||
| HCV/no HCV (n, %) | 17/121 (9.1/65.1) | 47 (25.3) | ||
M = male, F = female, ASAT = aspartate aminotransferase, ALAT = alanine aminotransferase, GGT = gamma-glutamyltransferase, AP = alkaline phosphatase, TBR = total bilirubin, HBV = hepatitis B infection, HCV = hepatitis C infection, IQR = interquartile range.
The population pharmacokinetics of ritonavir were best described by a one-compartment model with first-order absorption and elimination. Models with nonlinear Michaelis Menten elimination were investigated but proved to be less satisfactory than linear models with first-order elimination. Zero-order absorption and two compartmental models were studied but also turned out to be inadequate. However, the addition of an absorption lag-time (0.778 h), significantly improved the fit (ΔOFV = −150, P < 0.001). The residual error in ritonavir pharmacokinetics incorporated both an additive and a proportional component. The magnitude of the residual error was not constant across all individuals, which may influence parameter estimates. Therefore, several models were investigated to allow for interindividual varying residual error. Ultimately, inclusion of two different populations with different magnitudes of residual variability proved to be the most optimal model (ΔOFV = −48, P < 0.001). As a result, 64.8% of the population were associated with a relatively small additive error of 0.0600 mg l−1, whereas the remainder was associated with a larger additive error of 0.199 mg l−1. The proportional error for both populations was 15.4%. Including interoccasion variability in the apparent bioavailability in the model resulted in further optimization of the model.
The different covariates and the effects of indinavir, lopinavir and saquinavir on the pharmacokinetics of ritonavir were introduced separately in the model, using a univariate procedure. Only the introduction of lopinavir resulted in a statistically significant increase in goodness-of-fit, ΔOFV = −69.2 (P < 0.001) and a significant effect on the clearance of ritonavir. No other covariates were significantly related to the pharmacokinetics of ritonavir. The magnitude of increase in clearance was 272% during concomitant use of lopinavir. The following equation describes the final model for clearance:
in which LPV is 1 for individuals using lopinavir in their antiretroviral regimen and 0 for all others.
A correlation between the individual random effects of volume of distribution and absorption rate constant (ηV and ηka) was observed and covariance between these parameters was added to the model. The correlation coefficient was 0.868 (P = 0.001). In the final model the estimate of clearance was 10.5 l h−1 with an IIV of 38.3%. The estimates of volume of distribution and absorption rate constant were 96.6 l (IIV = 80.0%) and 0.871 h−1 (IIV = 169%), respectively. The calculated value for half-life from these estimates was 6.4 h. The results of the final pharmacokinetic model are summarized in Table 2.
Table 2.
Parameter estimates from the population pharmacokinetic model and the results of the bootstrap analysis
| Bootstrap analysis | ||||
|---|---|---|---|---|
| Parameter | Estimate | RSE (%) | Median | 95% PI |
| CL/F (l h−1) | 10.5 | 5.55 | 10.4 | 9.38–11.7 |
| θLPV | 2.72 | 11.7 | 2.72 | 2.20–3.67 |
| V/F (l) | 96.6 | 10.7 | 92.6 | 67.2–121 |
| ka (h−1) | 0.871 | 23.1 | 0.775 | 0.429–1.470 |
| Lag-time (h) | 0.778 | 4.91 | 0.768 | 0.388–0.867 |
| IIV CL/F (%) | 38.3 | 23.0 | 38.3 | 29.4–47.6 |
| IIV V/F (%) | 80.0 | 31.6 | 77.3 | 051.1–117.0 |
| IIV ka (%) | 169 | 25.8 | 163 | 113–209 |
| IOV F (%) | 59.1 | 15.6 | 58.5 | 48.4–67.6 |
| Correlation ηV-ηka | 0.868 | 34.7 | 0.876 | 0.718–1.000 |
| Fraction in P1 (%) | 64.8 | 18.5 | 63.5 | 37.2–84.1 |
| Additive error P1 (mg l−1) | 0.0600 | 13.5 | 0.0575 | 0.0353–0.1010 |
| Additive error P2 (mg l−1) | 0.199 | 15.2 | 0.202 | 0.0731–0.2760 |
| Proportional error (%) | 15.4 | 23.8 | 14.6 | 10.1–25.5 |
F = apparent bioavailability, CL/F = oral clearance, LPV = lopinavir, V/F = volume of distribution, ka = absorption rate constant, IIV = interindividual variability, IOV = interoccasion variability, P = population, RSE = residual standard error, PI = prediction interval.
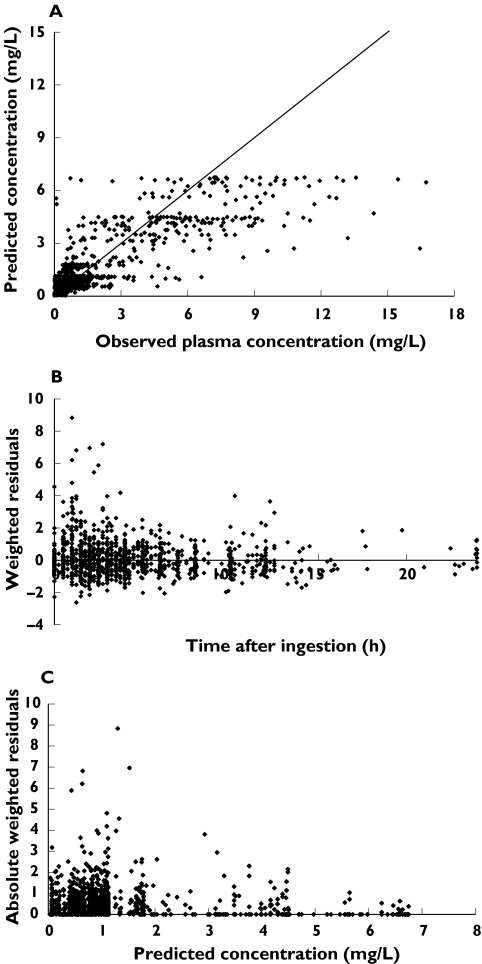
Figure 2 shows the model predicted plasma concentrations from the final model vs the observed concentrations of ritonavir, the weighted residuals vs time and the absolute weighted residuals vs predicted plasma concentrations.
Figure 2.
Model predicted concentrations vs observed concentrations of ritonavir (A), weighted residuals vs time (B) and absolute weighted residuals vs predicted concentrations of ritonavir (C) using the final model
From the original data set more than 1000 replicate bootstrap data sets were generated and used for the evaluation of the precision of the parameter estimates. Unsuccessful terminations due to boundaries were run again with enlarged borders. Abnormal and unsuccessful terminations were excluded from the bootstrap calculations. Successful minimizations and unsuccessful termination due to rounding errors with number of significance above 2 were included in the bootstrap calculations. However, less than 5% of the bootstrap runs were unsuccessful. Table 2 lists the results of the 1000 included bootstraps, presented as medians and 95% prediction intervals, and the parameter estimates of the final model with the corresponding relative standard error. Similar median bootstrap values to the parameter estimates of the original data set indicated acceptable precision.
Discussion
Despite the widespread use of ritonavir, few data are available on its pharmacokinetics in clinical practice. The aim of this study was to characterize the population pharmacokinetics of ritonavir and to identify any covariates. The model development started with a careful data check. The influence of outliers was studied extensively during this phase of model development. The combination of the described cut off of 0.01 mg l−1 with an additive residual error model as described proved to be the most efficient way to minimize the influence of possible nonadherence on the results of this study.
The pharmacokinetics of ritonavir were adequately described by a population pharmacokinetic model consisting of one compartment with first-order absorption with a lag-time and first-order elimination. This structural model was similar to that of Sale et al. [18]. However, Hsu et al. [17] described a pharmacokinetic model consisting of one compartment with first-order absorption and Michaelis-Menten saturable metabolism. In this study samples were collected between days 1 and 17. Thus only at the end of the study would subjects be at steady-state, which may have influenced the characterization of the pharmacokinetics of ritonavir.
Our estimation for volume of distribution fell within the wide range (28–123 l) of values found in previous studies [7, 17–19]. In addition, the estimates of clearance and half-life were similar to those found previously [7, 13, 17, 19]. Because both clearance (CL/F) and volume of distribution (V/F) depend on bioavailability (F), these pharmacokinetic parameters may be correlated. Large interindividual and interoccasion variability in clearance, volume of distribution, absorption rate constant and apparent bioavailability was observed (38.3%, 80.0%, 169% and 59.1%, respectively). Differences in protein binding, absorption, enzyme induction or variability in expression of CYP enzymes, Pgp or MRP1 may contribute to this variability, as may environmental factors and dietary habits. It has been reported that dose may be a determinant of the pharmacokinetics of ritonavir [17]. However, in the present work dosage was found to be a statistically nonsignificant and clinically nonrelevant covariate. Finally, all plasma samples were obtained after at least 14 days of treatment with ritonavir, suggesting that patients were at steady-state and thus that auto-induction of metabolism was no longer contributing to interoccasion variability.
Since data were obtained from different studies, it was anticipated that the residual error may not be constant across all subjects. Therefore, the individual contribution to the residual error was accounted for by including an interindividual variability term in the residual error. However, successful termination was not achieved. Therefore, two populations were introduced in the additive error, the reason for which could not be explained. Nevertheless, this approach to the residual variability resulted in less biased parameter estimates and improved goodness-of-fit of the model.
The concomitant use of lopinavir resulted in a 2.72 fold increase in the clearance of ritonavir, but saquinavir and indinavir were apparently without effect. It is known that trough plasma concentrations of ritonavir are significantly higher in patients receiving combinations containing saquinavir or indinavir than combinations with lopinavir or amprenavir [20, 21]. We had no patients on the combination amprenavir-ritonavir in our database, and thus the results of these studies [20, 21] may be compatible with our data.
The development of the current population pharmacokinetic model was undertaken for subsequent investigation of the pharmacokinetics of dual protease inhibitor regimens. Therefore, model validation was of particular importance. In the current study, the bootstrap resampling technique was performed as an internal validation. The 1000 replicate datasets yielded median model parameters that were comparable with the estimates of the original dataset, indicating the high precision of the developed model.
In conclusion, a model for the population pharmacokinetics of ritonavir was developed and validated. To this end, a large outpatient population was used, incorporating concentration-time points over the complete dosing interval. Except for the concomitant use of lopinavir, no patient characteristics influenced ritonavir pharmacokinetics. The model will be integrated into other population models to investigate the pharmacokinetics of other protease inhibitors used in combination with ritonavir, which may lead to a further optimization of ritonavir-containing antiretroviral therapy.
References
- 1.Gatti G, Di Biagio A, Casazza R, et al. The relationship between ritonavir plasma levels and side-effects: implications for therapeutic drug monitoring. AIDS. 1999;13:2083–9. doi: 10.1097/00002030-199910220-00011. [DOI] [PubMed] [Google Scholar]
- 2.Dumon C, Solas C, Thuret I, et al. Relationship between efficacy, tolerance, and plasma drug concentration of ritonavir in children with advanced HIV infection. Ther Drug Monit. 2000;22:402–8. doi: 10.1097/00007691-200008000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Danner SA, Carr A, Leonard JM, et al. A short-term study of the safety, pharmacokinetics, and efficacy of ritonavir, an inhibitor of HIV-1 protease. European-Australian Collaborative Ritonavir Study Group. N Engl J Med. 1995;333:1528–33. doi: 10.1056/NEJM199512073332303. [DOI] [PubMed] [Google Scholar]
- 4.Kempf DJ, Marsh KC, Kumar G, et al. Pharmacokinetic enhancement of inhibitors of the human immunodeficiency virus protease by co-administration with ritonavir. Antimicrob Agents Chemother. 1997;41:654–60. doi: 10.1128/aac.41.3.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Heeswijk RPG, Veldkamp AI, Mulder JW, et al. Once-daily dosing of saquinavir and low-dose ritonavir in HIV-1-infected individuals: a pharmacokinetic pilot study. AIDS. 2000;14:F103–F110. doi: 10.1097/00002030-200006160-00003. [DOI] [PubMed] [Google Scholar]
- 6.Goujard C, Vincent I, Meynard JL, et al. Steady-state pharmacokinetics of amprenavir co-administered with ritonavir in human immunodeficiency virus type 1-infected patients. Antimicrob Agents Chemother. 2003;47:118–23. doi: 10.1128/AAC.47.1.118-123.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hsu A, Granneman GR, Cao G, et al. Pharmacokinetic interaction between ritonavir and indinavir in healthy volunteers. Antimicrob Agents Chemother. 1998;42:2784–91. doi: 10.1128/aac.42.11.2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu A, Granneman GR, Cao G, et al. Pharmacokinetic interactions between two human immunodeficiency virus protease inhibitors, ritonavir and saquinavir. Clin Pharmacol Ther. 1998;63:453–64. doi: 10.1016/S0009-9236(98)90041-8. [DOI] [PubMed] [Google Scholar]
- 9.Kurowski M, Kaeser B, Sawyer A, Popescu M, Mrozikiewicz A. Low-dose ritonavir moderately enhances nelfinavir exposure. Clin Pharmacol Ther. 2002;72:123–32. doi: 10.1067/mcp.2002.126178. [DOI] [PubMed] [Google Scholar]
- 10.Kumar GN, Rodrigues AD, Buko AM, Denissen JF. Cytochrome P450-mediated metabolism of the HIV-1 protease inhibitor ritonavir (ABT-538) in human liver microsomes. J Pharmacol Exp Ther. 1996;277:423–31. [PubMed] [Google Scholar]
- 11.Von Moltke LL, Greenblatt DJ, Grassi JM, et al. Protease inhibitors as inhibitors of human cytochromes P450: high risk associated with ritonavir. J Clin Pharmacol. 1998;38:106–11. doi: 10.1002/j.1552-4604.1998.tb04398.x. [DOI] [PubMed] [Google Scholar]
- 12.Penzak SR, Hon YY, Lawhorn WD, Shirley KL, Spratlin V, Jann MW. Influence of ritonavir on olanzapine pharmacokinetics in healthy volunteers. J Clin Psychopharmacol. 2002;22:366–70. doi: 10.1097/00004714-200208000-00006. [DOI] [PubMed] [Google Scholar]
- 13.Hsu A, Granneman GR, Bertz RJ Ritonavir. Clinical pharmacokinetics and interactions with other anti-HIV agents. Clin Pharmacokinet. 1998;35:275–91. doi: 10.2165/00003088-199835040-00002. [DOI] [PubMed] [Google Scholar]
- 14.Huisman MT, Smit JW, Schinkel AH. Significance of P-glycoprotein for the pharmacology and clinical use of HIV protease inhibitors. AIDS. 2000;14:237–42. doi: 10.1097/00002030-200002180-00005. [DOI] [PubMed] [Google Scholar]
- 15.Olson DP, Scadden DT, D'Aquila RT, De Pasquale MP. The protease inhibitor ritonavir inhibits the functional activity of the multidrug resistance related-protein 1 (MRP-1) AIDS. 2002;16:1743–7. doi: 10.1097/00002030-200209060-00005. [DOI] [PubMed] [Google Scholar]
- 16.Molla A, Korneyeva M, Gao Q, et al. Ordered accumulation of mutations in HIV protease confers resistance to ritonavir. Nat Med. 1996;2:760–6. doi: 10.1038/nm0796-760. [DOI] [PubMed] [Google Scholar]
- 17.Hsu A, Granneman GR, Witt G, et al. Multiple-dose pharmacokinetics of ritonavir in human immunodeficiency virus-infected subjects. Antimicrob Agents Chemother. 1997;41:898–905. doi: 10.1128/aac.41.5.898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sale M, Sadler BM, Stein DS. Pharmacokinetic modeling and simulations of interaction of amprenavir and ritonavir. Antimicrob Agents Chemother. 2002;46:746–54. doi: 10.1128/AAC.46.3.746-754.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lu JF, Blaschke TF, Flexner C, Rosenkranz SL, Sheiner LB. Model-based analysis of the pharmacokinetic interactions between ritonavir, nelfinavir, and saquinavir after simultaneous and staggered oral administration. Drug Metab Dispos. 2002;30:1455–61. doi: 10.1124/dmd.30.12.1455. [DOI] [PubMed] [Google Scholar]
- 20.Lamotte C, Reynes J, Vildé JL, Yéni P, Katlama C, Peytavin G. 9th European Aids Conference (EACS), 1st EACS Resistance and Pharmacology Workshop. Poland: Warsaw; 2003. Ritonavir (RTV) trough plasma concentrations (Cmin) in RTV low dose boosted protease inhibitors (PI) containing regimen and their use for therapeutic drug monitoring in HIV-infected patients (pts) p. Abstract 4.3/4. 25–29 October. [Google Scholar]
- 21.Guiard-Schmid JB, Poirier JM, Meynard JL, et al. High variability of plasma drug concentrations in dual protease inhibitor regimens. Antimicrob Agents Chemother. 2003;47:986–90. doi: 10.1128/AAC.47.3.986-990.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brundage RC, Yong FH, Fenton T, Spector SA, Starr SE, Fletcher CV. Intrapatient variability of efavirenz concentrations as a predictor of virologic response to antiretroviral therapy. Antimicrob Agents Chemother. 2004;48:979–84. doi: 10.1128/AAC.48.3.979-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Heeswijk RPG, Veldkamp AI, Hoetelmans RMW, et al. The steady-state plasma pharmacokinetics of indinavir alone and in combination with a low dose of ritonavir in twice daily dosing regimens in HIV-1-infected individuals. AIDS. 1999;13:F95–F99. doi: 10.1097/00002030-199910010-00001. [DOI] [PubMed] [Google Scholar]
- 24.Veldkamp AI, van Heeswijk RPG, Mulder JW, et al. Steady-state pharmacokinetics of twice-daily dosing of saquinavir plus ritonavir in HIV-1-infected individuals. J Acquir Immune Defic Syndr. 2001;27:344–9. doi: 10.1097/00126334-200108010-00004. [DOI] [PubMed] [Google Scholar]
- 25.van Heeswijk RPG, Veldkamp AI, Mulder JW, et al. The steady-state pharmacokinetics of nevirapine during once daily and twice daily dosing in HIV-1-infected individuals. AIDS. 2000;14:F77–F82. doi: 10.1097/00002030-200005260-00001. [DOI] [PubMed] [Google Scholar]
- 26.Crommentuyn KML, Mulder JW, Mairuhu ATA, van Gorp ECM, Meenhorst PL, Huitema ADR, Beijnen JH. The plasma and intracellular steady-state pharmacokinetics of lopinavir/ritonavir in HIV-1-infected patients. Antivir Ther. 2004;9:779–785. [PubMed] [Google Scholar]
- 27.van Heeswijk RPG, Hoetelmans RMW, Harms R, et al. Simultaneous quantitative determination of the HIV protease inhibitors amprenavir, indinavir, nelfinavir, ritonavir and saquinavir in human plasma by ion-pair high-performance liquid chromatography with ultraviolet detection. J Chromatogr B Biomed Sci Appl. 1998;20:159–68. doi: 10.1016/s0378-4347(98)00392-2. [DOI] [PubMed] [Google Scholar]
- 28.Beal S, Sheiner L. NONMEM User's Guides. University of California at San Francisco: NONMEM Project Group; 1998. [Google Scholar]
- 29.Karlsson MO, Sheiner LB. The importance of modeling interoccasion variability in population pharmacokinetic analyses. J Pharmacokinet Biopharm. 1993;21:735–50. doi: 10.1007/BF01113502. [DOI] [PubMed] [Google Scholar]
- 30.Karlsson MO, Jonsson EN, Wiltse CG, Wade JR. Assumption testing in population pharmacokinetic models: illustrated with an analysis of moxonidine data from congestive heart failure patients. J Pharmacokinet Biopharm. 1998;26:207–46. doi: 10.1023/a:1020561807903. [DOI] [PubMed] [Google Scholar]
- 31.Parke J, Holford NH, Charles BG. A procedure for generating bootstrap samples for the validation of nonlinear mixed-effects population models. Comput Meth Programs Biomed. 1999;59:19–29. doi: 10.1016/s0169-2607(98)00098-4. [DOI] [PubMed] [Google Scholar]
- 32.Mould DR, Holford NH, Schellens JH, et al. Population pharmacokinetic and adverse event analysis of topotecan in patients with solid tumors. Clin Pharmacol Ther. 2002;71:334–48. doi: 10.1067/mcp.2002.123553. [DOI] [PubMed] [Google Scholar]