Abstract
Naproxen is a nonsteroidal anti-inflammatory drug widely used as an analgesic and anti-inflammatory agent. The conjugated forms of naproxen and O-DMN, its demethylated metabolite, account for 66–92% of naproxen found in human urine. In this study, O-DMN and structurally related compounds were tested as substrates for seven isoforms of human cytosolic sulfotransferase (SULT). The SULT2 or hydroxysteroid SULT isoforms, SULT2A1 and SULT2B1b, did not show reactivity with any of the compounds. All five SULT1 isoforms were active although there was variability between SULT isoforms and compounds assayed. O-DMN was sulphated by SULT1A1, SULT1B1 and SULT1E1. All five SULT1 isoforms were capable of conjugating both α-naphthol and β-naphthol. Apparent Km values for O-DMN sulphation were significantly higher than the values for either α-naphthol or β-naphthol. SULTs 1A1, 1B1 and 1E1 had Kms for O-DMN sulphation of 84 µm, 690 µm and 341 µm, respectively. These Km values were 40–1150-fold higher than the Km values for α- and β-naphthol. The role of the side-chain in O-DMN sulphation was studied using a series of structurally related β-naphthol compounds as substrates for SULT1A1 and SULT1E1. The presence of lipophilic groups increased affinity for both SULT isoforms whereas inclusion of a carboxyl group inhibited activity. These studies indicate that O-DMN is sulphated by SULT1A1, B1 and 1E1. Because of the high concentrations of SULT1A1 expression in human liver and intestines and its higher affinity for O-DMN sulphation, SULT1A1 may have a role in the first pass metabolism of O-DMN.
Keywords: Naproxen, drug metabolism, sulphation, sulfotransferase, naphthol
Introduction
Naproxen is a common nonsteroidal anti-inflammatory drug (NSAID) that has been approved for over-the-counter use since 1991 and is utilized for its short-term analgesic effects as well as in the treatment of certain types of arthritis, dysmennhorea and migraine attacks. The major Phase I metabolite of naproxen detected in man is 6-O-desmethylnaproxen (O-DMN) [1]. Naproxen undergoes Phase I dealkylation catalysed by the cytochrome P450s to the O-demethylated metabolite, and both compounds are biotransformed by Phase II metabolism into glucuronide conjugates as well as sulphated metabolites [2]. These conjugates are the primary urinary metabolites, with only a small amount of the drug being eliminated as either O-DMN or naproxen itself [2]. Approximately 95% of orally administered naproxen or metabolites is recovered in urine irrespective of the dose [1]. Various reports indicate that naproxen or O-DMN glucuronides constitute approximately 85%[3] while sulphoconjugates constitute approximately 11%[4] of the dose excreted. Although glucuronidation of both naproxen and O-DMN to their acyl glucuronides has been well studied, there is little information available as to the sulphoconjugation these compounds. Naproxen itself is not a candidate for sulphation due to the absence of a free hydroxyl group, however, sulphation of O-DMN has been reported [4]. Analysis of the SULT isoforms involved in O-DMN metabolism will provide insights into the first-pass and tissue specific metabolism of naproxen.
Sulphation is one of the major conjugation reactions involved in Phase II drug metabolism. The cytosolic sulphotransferase (SULT) gene family is responsible for the sulphation of many endogenous and xenobiotic compounds such as steroids, bile acids, hormones and neurotransmitters as well as many drugs and drug metabolites. SULTs catalyse the transfer of the sulphonate group from 3′-phosphoadenosine-5′-phosphosulphate (PAPS) to the hydroxyl or amino group of various compounds to form either a sulphate or sulphamate [5]. Sulphate conjugation of most classes of compounds renders them more hydrophilic so as to facilitate their excretion. In addition, the presence of the charged sulphate moiety decreases or abolishes the biological activity of most compounds.
The SULT1 or phenolic SULT gene family consists of at least seven distinct isoforms. The activity of five members of the SULT1 family with O-DMN and related compounds was investigated in this study; these include SULT1A1, SULT1A3, SULT1B1, SULT1C2 and SULT1E1. SULT1A1 catalyses the sulphation of many small neutral phenols and is expressed in many tissues including liver, small intestine and platelets [5–8]. SULT1A3 has a high affinity for the sulphation of monoamine neurotransmitters and is also capable of sulphating many small phenols [9, 10]. SULT1A3 is expressed in platelets and small intestine; however, it is expressed at much lower concentrations in liver than SULT1A1 [6, 11]. SULT1B1 and SULT1C2 also sulphate small phenols and are expressed at low concentrations in intestine [12–14]. SULT1E1 is responsible for the high affinity conjugation of β-oestradiol and is expressed at low concentrations in liver and intestine [15, 16]. SULT1E1 also sulphates many small phenols but with affinities at least 500–1000-fold less than that for β-oestradiol [15–17].
The SULT2 or hydroxysteroid SULT gene family consists of two genes, SULT2A and SULT2B. SULT2A1 is highly expressed in adrenal and liver and conjugates 3α- and 3β-hydroxysteroids as well as the aliphatic hydroxyls of xenobiotics [18–20]. The SULT2B1 gene encodes two isoforms, SULT2B1a and SULT2B1b, which result from the use of alternate transcriptional start sites and the incorporation of different initial exons [21]. Although both SULT2B1a and SULT2B1b messages are present in human tissues, only SULT2B1b protein expression has been detected in human tissues [22, 23]. Both expressed SULT2B1 isoforms are highly selective for the sulphation of 3β-hydroxysteroids although several xenobiotics are also substrates [22].
In this study, the ability of seven isoforms of human SULT to sulphate O-DMN was evaluated. Additionally, O-DMN structural analogues were utilized to investigate the role of the different structural features in regulating sulphation. Following an initial analysis to determine which SULT isoforms were active in sulphating the different compounds, the kinetic properties of the SULT isoforms involved were characterized. The role of O-DMN structural alterations in regulating sulphation was then evaluated through comparison of the kinetic parameters for the major SULTs involved in O-DMN sulphation.
Methods
Materials
[35S]3′-Phosphoadenosine 5′-phosphosulphate (PAPS) (3 Ci mmol−1) was purchased from Perkin Elmer Life Sciences (Boston, MA). Non-radiolabelled PAPS was purchased from Dr Sanford Singer (University of Dayton, Dayton, OH). LK6DF 60 Å silica gel thin layer chromatography (TLC) plates with a layer thickness of 250 µm were obtained from Whatman Inc. (Clifton, NJ). α-Naphthol and β-naphthol were purchased from Sigma Chemical Co. (St. Louis, MO). All other chemicals used were of reagent grade from Fisher Scientific (Norcross, GA).
Synthesis of structurally related naproxen analogues
O-DMN was prepared by mixing S-naproxen (10.6 mmol) in hydrobromic acid (30%), acetic acid and water for 5 h at 105 °C. The reaction mixture was poured onto 50 g of ice-water and the precipitate was filtered off. The crude product was purified on a flash column using heptane : ethyl acetate (1 : 1) as solvent. The pure fractions were recrystallized from heptane : ethyl acetate (1 : 5) to give 0.6 g of demethylated naproxen.
6-Methyl-naphthalen-2-ol was synthesized by adding dry tetrahydrofuran (THF) (4 mL) to 2.0 mmol of 2-bromo-6-methoxy-naphthalen at −78 °C. N-butyl lithium (2.4 mmol) was added drop-wise to the solution followed by methyl iodide (2.1 mmol) after a 1-h incubation. The reaction was allowed to warm to room temperature and quenched with 1 m HCl. After extraction with ethyl acetate 350 mg of crude material was recovered. The crude fraction was demethylated in 5 mL dichloromethane by adding 1 eq of 1.0 m BBr3 at 0 °C followed by a 2-h incubation at ambient temperature. The reaction was quenched with 10 mL of brine and the organic layer was dried (Na2SO4) and evaporated. Flash column purification was carried out with heptane : ethyl acetate (1 : 1) as solvent. The product was re-crystallized from 2 mL heptane : ethyl acetate (1 : 5) and yielded 200 mg of pure product.
Similarly, 6-ethyl-naphthalen-2-ol was prepared by adding dry THF (4 mL) to 2.0 mmol of 2-bromo-6-methoxy-naphthalen at −78 °C. N-butyl lithium (2.4 mmol) was added drop-wise to the above solution. After 1 h, 0.12 mL (2.1 mmol) of ethyl iodide was added and the reaction mixture was allowed to warm up to room temperature then quenched with 5 mL of 1 m HCl. Extraction with ethyl acetate recovered 373 mg of crude material. The crude fraction was demethylated in 5 mL dichloromethane by adding 1 eq of 1.0 m BBr3 at 0 °C and then incubated at ambient temperature for 2 h. The reaction was quenched with 10 mL of brine and the organic layer was dried (Na2SO4) and evaporated. Purification on a flash column with heptane : ethyl acetate (1 : 1) as solvent gave 300 mg of 6-ethyl-naphthalen-2-ol. Recrystallization from 3 mL heptane gave 230 mg of pure product.
6-hydroxy-naphthalen-2-carboxylic acid was synthesized by adding dry THF (5 mL) to 1.43 g (6.0 mmol) 2-bromo-6 methoxy-naphthalen. N-butyl lithium (4.2 mL, 6.6 mmol) was added at −78 °C. After 1 h solid CO2 was added and the solution stirred for 1 h. The reaction mixture was then poured onto ice-water; following base/acid extraction, 950 mg of the carboxylic acid was recovered. The carboxylic acid (3.06 mmol) was dissolved in 15 mL of dichloromethane then 12.2 mL (12.2 mmol) of BBr3 was added at 0 °C. The mixture was stirred for 1 h at room temperature and quenched with 20 mL of brine. The organic layer was separated, dried (Na2SO4) and evaporated. Flash chromatography with heptane : ethyl acetate (1 : 1) as solvent gave 420 mg of 6-hydroxy-naphthalen-2-carboxylic acid.
To synthesize 6-hydroxmethyl-naphthalen-2-ol, 220 mg of 6-hydroxy-naphthalen-2-carboxylic acid was stirred with four equivalents of lithium aluminium hydride in 5 mL THF at room temperature for 5 h. The reaction mixture was quenched with 20 mL brine and extracted twice with ethyl acetate. Flash chromatography with ethyl acetate : methanol : NH3(c) (100 : 10 : 1) gave 130 mg of pure product. The structures of O-DMN and related compounds are shown in Figure 1.
Figure 1.
Structure of O-DMN and structurally related compounds
Sulphation assays
Sulphation activity was determined using O-DMN and related compounds as substrates with each of the bacterially expressed human SULT isoforms. The bacterial expression and purification of the human expressed SULTs, has been previously reported [7, 10, 12, 15, 24]. Briefly, the SULTs were expressed in E. coli using the pKK233-2 vector to generate the native form of the enzymes and purified by DEAE-Sepharose chromatography to obtain a preparation suitable for enzymatic characterization [25]. Sulphation assays using O-DMN and its structural analogues as substrates were performed with each of the expressed human SULTs using the assay procedure for nonradiolabelled steroids [12, 15]. Sulphation is assayed using the appropriate substrate and [35S]-PAPS as the sulphate donor with the subsequent resolution of [35S]-sulphated products by TLC [15, 22]. SULT1A1 reactions contained the appropriate substrate dissolved in ethanol, 50 mm Tris-HCl, pH 7.4 and 5 µm PAPS in a final volume of 62.5 µL; all other SULT reactions were supplemented with the addition of 10 mm MgCl2. Control reactions were run with no substrate but contained the appropriate volume of the ethanol vehicle. Reactions were incubated for appropriate times at 37 °C then terminated by spotting a 50-µL aliquot of each reaction on a silica gel TLC plate. The plate was developed in methylene chloride : MeOH : ammonium hydroxide (85 : 15 : 5 by volume) and the radiolabelled sulphated products were localized by autoradiography. The sulphated products were scraped into scintillation fluid and radioactivity was determined by scintillation spectroscopy [8, 22].
Initially, substrate concentration curves were performed to determine the effective concentration range for sulphation. For Km and Vmax calculations, and particularly to avoid the effects of substrate inhibition observed with many compounds and the SULT isoforms, varying concentrations of substrate were added within the established linear range of sulphated product formation. The range of substrate concentrations for analysis of kinetic properties was generally 0.13–2 µm. Kinetic experiments were analysed by using Enzyme Kinetics software (Trinity Software, Campton, NH).
Results
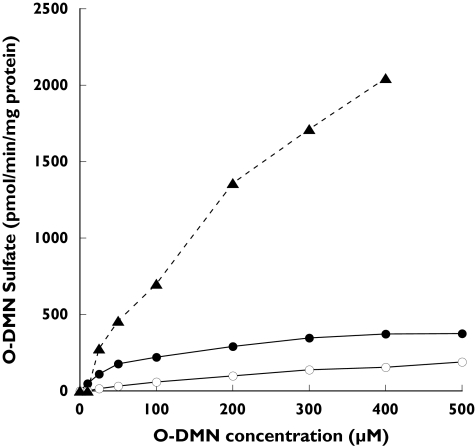
To ascertain the ability of seven major human SULT isoforms to sulphate O-DMN, reactions were run using O-DMN as substrate with the expressed human SULTs. The expressed and purified human SULTs were assayed with O-DMN as substrate at concentrations from 0 to 500 µm. Of the SULT isoforms tested only SULT1A1, SULT1B1, and SULT1E1 utilized O-DMN as a substrate. Figure 2 shows that O-DMN-sulphate formation catalysed by the three SULTs increased with increasing O-DMN concentrations. Substrate inhibition was not observed with O-DMN concentrations up to 500 µm. As shown in Table 1, SULT1A1 had the lowest Km (84 µm) for the sulphation of O-DMN although the sulphation rate reached a plateau at concentrations above 200 µm. SULT1E1 had a lower affinity for O-DMN although its sulphation activity increased steadily up to a concentration of 400 µm. SULT1B1 displayed trace concentrations of activity that increased in a linear manner up to an O-DMN concentration of 500 µm.
Figure 2.
Concentration curves for sulphation of O-DMN by SULT1A1 (•), SULT1B1 (○), and SULT1E1 (▴). Reactions were run and [35S]-labelled products analysed by TLC as a described in Methods. O-DMN concentrations ranged from 0 to 500 µM
Table 1. Sulphation of O-DMN by human cytosolic SULT1 isoforms.
| Substrate | SULT1A1 | SULT1A3 | SULT1B1 | SULT1C2 | SULT1E1 |
|---|---|---|---|---|---|
| O-DMN | 84 ± 6 (8.4) | ND | 690 ± 131 (69.0) | ND | 341 ± 51 (26.5) |
| α-Naphthol | 2.1 ± 1.3 (247) | 4.8 ± 0.4 (201) | 0.6 ± 0.1 (2586) | 1.0 ± 0 (65 622) | 2.4 ± 0.2 (221) |
| β-Naphthol | 0.4 ± 0 (120) | 116 ± 11.3 (112) | 1.5 ± 0.5 (735) | 0.4 ± 0 (60 133) | 3.0 ± 0.4 (307) |
Apparent Km values represent the mean of three separate determinations ± standard deviation. Km values were determined using the Enzyme Kinetics program (Trinity Software). ‘ND’ indicates no detectable activity at the substrate concentrations tested. The Vmax values in pmol min−1 mg−1 are in parentheses next to the Km values.
O-DMN has a dual conjugated ring structure based on the structure of β-naphthol (Figure 1). To understand how the position of the hydroxyl group affects SULT and substrate affinity and activity, the ability of the human SULTs to sulphate α- and β-naphthol in comparison to O-DMN was investigated. All five of the SULT1 family members tested sulphated both α- and β-naphthol (Table 1) whereas neither of the SULT2 isoforms showed activity with these substrates. All five of the SULT1 isoforms sulphated α-naphthol with Km values between 0.6 and 4.8 µm. SULT1B1 displayed the lowest Km (0.6 µm) and SULT1A3 the highest (4.8 µm). The sulphation of β-naphthol by the SULT1 isoforms was more variable. SULT1A1 and SULT1C2 showed the lowest Kms (0.4 µm) for β-naphthol conjugation (Table 1). In contrast, SULT1A3 had a 24-fold higher Km for the sulphation of β-naphthol as compared with α-naphthol. However, the Km values of SULT1A1 and SULT1C2 for β-naphthol decreased 5 and 2.5-fold, respectively, as compared with those for α-naphthol. Therefore, the position of the hydroxyl group on the naphthol ring system significantly altered the binding of SULT1A3 as compared with the other SULT1 isoforms.
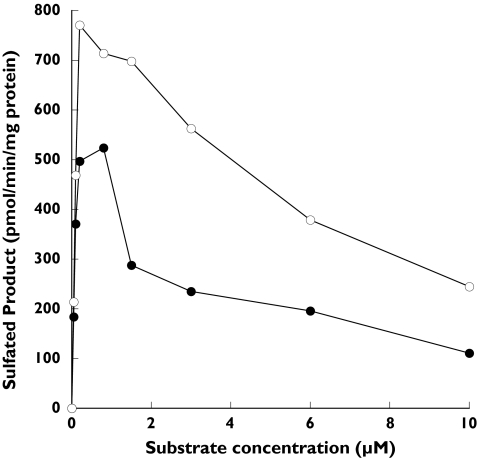
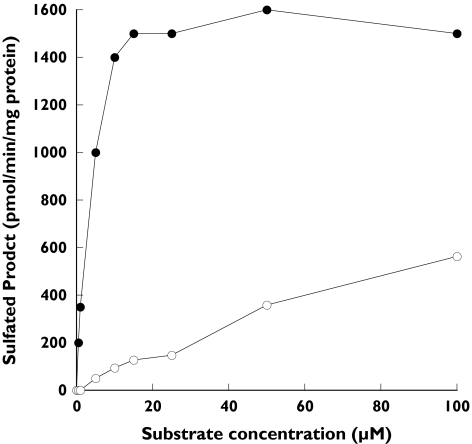
SULT1A1 and SULT1A3 are 93% identical in amino acid sequence and believed to be derived by gene duplication. Although the SULT1A isoforms are very similar they display significant differences in substrate interactions as demonstrated by their very different Km values for β-naphthol sulphation. SULT1A1 demonstrated substrate inhibition with both α- and β-naphthol that did not occur with the O-DMN substrate (Figure 3). Maximal activity was observed with both α- and β-naphthol substrates at concentrations below 1 µm whereas maximal sulphation of O-DMN occurred at much higher concentrations in the range of 300 µm (Figure 2). In contrast, Figure 4 demonstrates that substrate inhibition was not observed using SULT1A3 with concentrations of either α- or β-naphthol up to 100 µm. There was no detectable sulphation of O-DMN by SULT1A3 (Table 1). Additionally, the difference in the sulphation of α- and β-naphthol rates by SULT1A3 is apparent in Figure 4. SULT1E1, which shows a slightly lower affinity for α-naphthol sulphation than SULT1A1 (Table 1), also demonstrated substrate inhibition with maximal activity at approximately 5 µm for both α- and β-naphthol (data not shown).
Figure 3.
Concentration curves for sulphation of α-naphthol (•) and β-naphthol (○) by SULT1A1. Reactions were run and [35S]-labelled products analysed by TLC as described in Methods with either α- or β-naphthol as substrates at concentrations between 0 and 10 µM
Figure 4.
Concentration curves for sulphation of α-naphthol (•) and β-naphthol (○) by SULT1A3. Reactions were run and [35S]-labelled products analysed by TLC as described in Methods with either α- or β-naphthol as substrates at concentrations between 0 and 100 µM
Because the structure of O-DMN is based upon that of β-naphthol with the addition of a side chain at the C6 of the B-ring (Figure 1), structural analogues of O-DMN with alterations in this side chain were synthesized to investigate their effects on sulphation activity. Since the position of the hydroxyl group in the naphthol structure significantly affected only the activity of SULT1A3, the role of the side-chain in altering sulphation activity of the β-naphthol hydroxyl group was investigated utilizing SULT1A1 and 1E1, the two SULTs with the highest affinity for the sulphation of O-DMN. Table 2 summarizes the kinetic data for sulphation of the structural analogues of O-DMN by SULT1A1. The addition of methyl and ethyl groups resulted in stepwise increases in substrate affinity by SULT1A1 up to a 10-fold greater affinity as compared with β-naphthol. The addition of a hydroxymethyl group at the C6 resulted in a 2-fold decrease in affinity and had little effect on the affinity of the compound as compared with 6-methyl-naphthalen-2-ol. The insertion of a carboxylic acid group at the C6 position abolished sulphation activity. Substrate inhibition was observed with SULT1A1 using all three structural analogues (Table 2). The concentrations of the individual compounds generating maximal activity were in the same order rank as the Km values. The calculated Vmax values of the sulphation of the three structural analogues by SULT1A1 were not significantly different.
Table 2. Sulphation of structural analogues of O-DMN by SULT1A1.
| Substrate | Km (µM) | Vmax (pmol min−1 mg−1 protein) | Substrate inhibition |
|---|---|---|---|
| 6-Methyl-naphthalen-2-ol | 0.141 ± 0.02 | 240 ± 140 | Yes |
| 6-Ethyl-naphthalen-2-ol | 0.043 ± 0.03 | 202 ± 103 | Yes |
| 6-Hydroxy-naphthalen-2-carboxylic acid | ND | ND | NA |
| 6-Hydroxymethyl-naphthalen-2-ol | 0.318 ± 0.01 | 190 ± 39 | Yes |
Apparent Km values represent the mean of three separate determinations ± standard deviation. Km values were determined using the Enzyme Kinetics program (Trinity Software). ‘ND’ indicates no detectable activity at the substrate concentrations tested. ‘NA’ indicates ‘not applicable’.
Table 3 summarizes the kinetic data for sulphation of the O-DMN structural analogues by SULT1E1. The addition of a methyl or ethyl group at the C6 position decreased the Km values in a step-wise manner similar to what was observed with SULT1A1. In contrast to SULT1A1, the presence of the hydroxymethyl group in the ring structure increased the apparent Km values greater than 5-fold as compared with 6-methyl-naphthalen-2-ol. Similar to SULT1A1, incorporation of the carboxylic acid moiety abolished sulphation activity. Substrate inhibition was observed with 6-ethyl-naphthalen-2-ol and 6-methyl-naphthalen-2-ol at concentrations above 2 µm and 15 µm, respectively. Substrate inhibition was not observed with 6-hydroxymethyl-naphthalen-2-ol as a substrate up to a concentration of 100 µm.
Table 3. Sulphation of structural analogues of O-DMN by SULT1E1.
| Substrate | Km (µM) | Vmax (pmol min−1 mg−1 protein) | Substrate inhibition |
|---|---|---|---|
| 6-Methyl-naphthalen-2-ol | 1.3 ± 0.4 | 267 ± 68 | Yes |
| 6-Ethyl-naphthalen-2-ol | 0.7 ± 0.4 | 338 ± 197 | Yes |
| 6-Hydroxy-naphthalen-2-carboxylic acid | ND | ND | NA |
| 6-Hydroxymethyl-naphthalen-2-ol | 7.0 ± 1.2 | 217 ± 16 | No |
Apparent Km values represent the mean of three separate determinations ± standard deviation. Km values were determined using the Enzyme Kinetics program (Trinity Software). ‘ND’ indicates no detectable activity at the substrate concentrations tested. ‘NA’ indicates ′not applicable'.
Discussion
Although the pharmacological properties and therapeutic utility of naproxen have been extensively evaluated, there are aspects of naproxen metabolism that have not been completely elucidated. O-DMN is the major Phase I metabolite of naproxen [1] but is comparatively inactive with less than 1% of the anti-inflammatory potency of naproxen. Naproxen undergoes dealkylation to O-DMN primarily by CYP1A2 and CYP2C9 activity in human liver followed by Phase II conjugation [26]. Hence, naproxen itself may be glucuronidated to form naproxen acylglucuronide or isoglucuronide, or oxidized to O-DMN and conjugated to form either 6-O-DMN glucuronide [26] or 6-O-DMN sulphate [4]. The glucuronides of naproxen itself may comprise approximately 60% of the excreted dose while O-DMN glucuronide conjugates and sulphoconjugates comprise 25% and 11% of this dose, respectively [4]. Whether O-DMN is preferentially glucuronidated or sulphated to facilitate elimination may also be a tissue-specific effect dependent upon the enzymatic profile of the tissue.
In this study, we have demonstrated that SULT1A1, SULT1B1 and SULT1E1 are capable of conjugating O-DMN. SULT1A1 is the major xenobiotic phenol-sulphating isoform and was the most effective SULT isoform in conjugating O-DMN with a Km of 84 µm. SULT1B1 and SULT1E1 had higher Km values for O-DMN sulphation (690 and 341 µm, respectively) and thus may not play as great a physiological role in O-DMN metabolism. Additionally, SULT1A1 is the most highly expressed SULT1 isoform in liver implying that it is responsible for the majority of O-DMN sulphation especially following absorption in the GI tract. Franssen et al. [27] have suggested that the kidneys are the major site of naproxen demethylation and sulphation. However, both CYP2C9 and SULT1A1 are widely expressed in human tissues and at high concentrations in liver and GI tract suggesting that demethylation and conjugation occur in the liver [6, 28]. Also, impaired renal function had no effect on the pharmacokinetic parameters of naproxen [29] suggesting that demethylation and conjugation occur before naproxen is transported to the kidney. However, renal function may play an indirect role in O-DMN sulphation because O-DMN sulphate is present at elevated concentrations in the plasma of patients with impaired renal function, and the extent of sulphation is directly related to the severity of the renal dysfunction [4].
Structurally, O-DMN contains a β-naphthol backbone (Figure 1); however, it is a much poorer substrate for sulphation than α- or β-naphthol. To better understand the sulphation of O-DMN, the reactivity of the SULT1 isoforms with α- and β-naphthol was analysed to determine the effect of the orientation of the hydroxyl group on sulphation activity. All five SULT1 isoforms conjugated both α- and β-naphthol. The kinetic parameters for the sulphation of α- and β-naphthol by the SULT1 isoforms were similar with the exception of SULT1A3. SULT1A3 has a relatively high affinity for the conjugation of monoamine neurotransmitters such as dopamine and a lower affinity for the conjugation of many small phenols. SULT1A1 and SULT1C2 had the lowest Km values for the sulphation of α- and β-naphthol with some selectivity for β-naphthol (Table 1). In contrast, SULT1A1 was able to sulphate O-DMN whereas no activity was detected with SULT1C2.
Because all of the SULT1 isoforms have very different reactivities towards O-DMN as compared with β-naphthol, the 6-side-chain is important in determining SULT activity. To evaluate the effect of the 6-side-chain, structural analogues of O-DMN based upon β-naphthol were synthesized. The ability of these compounds to be sulphated by SULT1A1 and 1E1, the two isoforms with the highest affinities for O-DMN sulphation, was investigated. Sulphation of the structural analogues indicates that adding hydrophobic methyl or ethyl groups increases the affinity for binding in the active site. For both SULT1A1 and SULT1E1 there was a stepwise decrease in the Km value with the addition of the methyl then ethyl group. The incremental changes in the binding energy (ΔΔG) of the β-naphthol derivatives were also calculated for both SULT1A1 and SULT1E1 (Table 4). Addition of the methyl and ethyl groups to β-naphthol results in step-wise increases in the energetics of specificity of binding as indicated by negative ΔΔG-values [30]. Also, for both SULT isoforms the addition of the hydroxyl moiety to the methyl group of 6-methyl-naphthalen-2-ol results in a decrease in the stability of the interaction of the substrate with the enzymes. When comparing these ΔΔG-values with those obtained when similar moieties are moved from n-octanol to water [30], both sets of data are in the same range. This suggests that the net gain or loss of energy for these substrates and enzymes depends only on the net entropy gained from water molecules released from the substrate and enzyme when the enzyme binds to the substrate. An exception seems to be the smaller loss in energy than expected for 6-OH-methyl-naphtalen-2-ol compared with 6-methyl-naphtalen-2-ol for SULT1A1. However, in general both SULT1A1 and SULT1E1 show no unexplained preference for the investigated substrates, based on the ΔΔG-values, which can be expected for this class of broad substrate specificity metabolizing enzymes. Inspection of the model structure of SULT1A1 proposed by Barnett et al. [31] suggests that addition of the methyl and ethyl groups increases interaction with the hydrophobic residues V148 and F247 in the active site. Addition of a hydroxymethyl group to the β-naphthol ring system increases the Km values for both SULT1A1 and SULT1E1. This would be consistent with the disruption of the hydrophobic interactions stabilizing the binding of the methyl group.
Table 4. Free energy of binding values (ΔΔG) for sulphation structural analogues of O-DMN by SULT1A1 and SULT1E1.
| Joules mol−1 | ||
|---|---|---|
| Reaction | SULT1A1 | SULT1E1 |
| β-Naphthol → 6-Methyl-naphthalen-2-ol | −4475 | −1787 |
| 6-Methyl-naphthalen-2-ol → 6-Ethyl-naphthalen-2-ol | −2618 | −2203 |
| 6-Methyl-naphthalen-2-ol → 6-OH-methyl-naphthalen-2-ol | +2714 | +4875 |
The incremental changes in the binding energy (ΔΔG) of the β-naphthol derivatives were calculated for both SULT1A1 and SULT1E1 using the equation ΔΔG = -RTln(Vmax/Km)sub1/((Vmax/Km)sub2 [30]. A negative sign represents an increase in the stability of the interaction.
In contrast to the O-DMN structural analogues containing methyl or ethyl groups, no activity was observed with either SULT1A1 or SULT1E1 using 6-hydroxy-naphthalene-2-carboxylic acid as substrate (Figure 1). Thus, addition of this carboxylic acid group effectively abolishes the sulphation activity that these two SULTs possess with β-naphthol as substrate. However, these SULTs are both active with O-DMN, which differs from this 6-hydroxy-naphthalene-2-carboxylic acid substrate only by the addition of an additional carbon in the side-chain. It is apparent that this carbon in the carboxylic acid side-chain is critical to SULT activity, either by virtue of side-chain length or by effects on hydrophobicity involved in stabilization of the side-chain in the active site. A longer side-chain could potentially remove steric interference by the carboxylic acid group in the active site. Thus, in contrast to 6-hydroxy-naphthalene-2-carboxylic acid, O-DMN, which has a carboxylic acid group physically more removed from the molecular ring system, is sulphated by both SULT1A1 and SULT1E1 although the Km values are relatively high compared with β-naphthol and the methyl- and ethyl-substituted compounds.
Substrate inhibition is a common feature of the conjugation of high affinity substrates by the SULT isoforms [5, 25] and is generally associated with allosteric modulation of catalytic activity by substrate binding to an allosteric site separate from the catalytic site [17, 31, 32]. Both α- and β-naphthol showed substrate inhibition with SULT1A1 (Figure 3) whereas no inhibition was observed with SULT1A3 even though α-naphthol is a relatively high affinity substrate (Figure 4), indicating that alllosteric modulation may not occur in this case. Substrate inhibition was also observed during sulphation of the β-naphthol structural analogues by SULT1A1 (Table 2) and SULT1E1 (Table 3) with exception of 6-hydroxymethyl-naphthalen-2-ol sulphation by SULT1E1. 6-Hydroxymethyl-naphthalen-2-ol is a relatively low affinity substrate for SULT1E1 so the lack of substrate inhibition is not unanticipated. Neither SULT1A1, SULT1B1 nor SULT1E1 showed substrate inhibition during the sulphation of O-DMN (Figure 2). Again, the lack of substrate inhibition during the sulphation of O-DMN is consistent with its relatively low affinity for the SULT isoforms.
As demonstrated by this study, there are three human SULTs that conjugate O-DMN and may play a significant role in its metabolism and subsequent elimination. The presence of SULT1A1, SULT1B1 and SULT1E1 in intestine and high concentrations of SULT1A1 in human liver suggest that sulphation may have a role in the first pass metabolism of O-DMN. The greater affinity of SULT1A1 for O-DMN sulphation as well as its abundance in human liver suggest it is the primary SULT isoform involved. The relatively low affinity of O-DMN as a substrate is associated with the presence of the carboxyl group since deletion or substitution of this moiety leads to lower Km values.
Acknowledgments
The authors would like to thank Michelle Smith-Johnson for expert technical assistance and Josie L. Falany for reading the manuscript and helpful comments. This work was supported in part by a grant from AstraZeneca to CNF and by NIH grant GM38953 to.
References
- 1.Segre EJ. Naproxen metabolism in man. J Clin Pharmacol. 1975;15:316–23. doi: 10.1002/j.1552-4604.1975.tb01458.x. [DOI] [PubMed] [Google Scholar]
- 2.Davies NM, Anderson KE. Clinical pharmacokinetics of naproxen. Clin Pharmacokinet. 1997;32:268–93. doi: 10.2165/00003088-199732040-00002. [DOI] [PubMed] [Google Scholar]
- 3.Vree TB, van den Biggelaar-Martea M, Verwey-van Wissen CP. Determination of naproxen and its metabolite O-desmethylnaproxen with their acyl glucuronides in human plasma and urine by means of direct gradient high-performance liquid chromatography. J Chromatogr. 1992;578:239–49. doi: 10.1016/0378-4347(92)80422-m. [DOI] [PubMed] [Google Scholar]
- 4.Kiang CH, Lee C, Kushinsky S. Isolation and identification of 6-desmethylnaproxen sulfate as a new metabolite of naproxen in human plasma. Drug Metab Dispos. 1989;17:43–8. [PubMed] [Google Scholar]
- 5.Falany CN. Enzymology of human cytosolic sulfotransferases. FASEB J. 1997;11:206–16. doi: 10.1096/fasebj.11.4.9068609. [DOI] [PubMed] [Google Scholar]
- 6.Falany CN, Vazquez ME, Heroux JA, Roth JA. Purification and characterization of human liver phenol-sulfating phenol sulfotransferase. Arch Biochem Biophys. 1990;278:312–8. doi: 10.1016/0003-9861(90)90265-z. [DOI] [PubMed] [Google Scholar]
- 7.Wilborn TW, Comer KA, Dooley TP, Reardon IM, Heinrikson RL, Falany CN. Sequence analysis and expression of the cDNA for the phenol-sulfating form of human liver phenol sulfotransferase. Mol Pharmacol. 1993;43:70–7. [PubMed] [Google Scholar]
- 8.Falany CN, Wheeler J, Oh TS, Falany JL. Steroid sulfation by expressed human cytosolic sulfotransferases. J Steroid Biochem Mol Biol. 1994;48:369–75. doi: 10.1016/0960-0760(94)90077-9. [DOI] [PubMed] [Google Scholar]
- 9.Reiter C, Mwaluko G, Dunnette J, Van Loon J, Weinshilboum R. Thermolabile and thermostable human platelet phenol sulfotransferase: Substrate specificity and physical separation. Naunyn-Schmiedeberg's Arch Pharmacol. 1983;324:40–1147. doi: 10.1007/BF00497020. [DOI] [PubMed] [Google Scholar]
- 10.Ganguly TC, Krasnykh V, Falany CN. Bacterial expression and kinetic characterization of the human monoamine-sulfating form of phenol sulfotransferase. Drug Metab Dispos. 1995;23:945–50. [PubMed] [Google Scholar]
- 11.Weinshilboum RM. Phenol sulfotransferase in humans: properties, regulation, and function. Federation Proceedings. 1986;45:2223–8. [PubMed] [Google Scholar]
- 12.Wang J, Falany JL, Falany CN. Expression and characterization of a novel thyroid hormone-sulfating form of cytosolic sulfotransferase from human liver. Mol Pharmacol. 1998;53:274–82. doi: 10.1124/mol.53.2.274. [DOI] [PubMed] [Google Scholar]
- 13.Freimuth RR, Raftogianis RB, Wood TC, Moon E, Kim UJ, Xu J, Siciliano MJ, Weinshilboum RM. Human sulfotransferases SULT1C1 and SULT1C2: cDNA characterization, gene cloning, and chromosomal localization. Genomics. 2000;65:157–65. doi: 10.1006/geno.2000.6150. [DOI] [PubMed] [Google Scholar]
- 14.Freimuth RR, Eckloff B, Wieben ED, Weinshilboum RM. Human sulfotransferase SULT1C1 pharmacogenetics: gene resequencing and functional genomic studies. Pharmacogenetics. 2001;11:747–56. doi: 10.1097/00008571-200112000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Falany CN, Krasnykh V, Falany JL. Bacterial expression and characterization of a cDNA for human liver estrogen sulfotransferase. J Steroid Biochem Mol Biol. 1995;52:529–39. doi: 10.1016/0960-0760(95)00015-r. [DOI] [PubMed] [Google Scholar]
- 16.Her C, Szumlanski C, Aksoy I, Weinshilboum R. Human jejunal estrogen sulfotransferase and dehydroepiandrosterone sulfotransferase: immunological characterization of individual variation. Drug Metabolism Disposition. 1996;24:1328–35. [PubMed] [Google Scholar]
- 17.Zhang H, Varlamova O, Vargas FM, Falany CN, Leyh TS, Varmalova O. Sulfuryl transfer: the catalytic mechanism of human estrogen sulfotransferase. J Biol Chem. 1998;273:10888–92. doi: 10.1074/jbc.273.18.10888. [DOI] [PubMed] [Google Scholar]
- 18.Falany CN, Vazquez ME, Kalb JM. Purification and characterization of human liver dehydroepiandrosterone sulphotransferase. Biochem J. 1989;260:641–6. doi: 10.1042/bj2600641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Comer KA, Falany CN. Immunological characterization of dehydroepiandrosterone sulfotransferase from human liver and adrenal. Mol Pharmacol. 1992;41:645–51. [PubMed] [Google Scholar]
- 20.Falany CN, Comer KA, Dooley TP, Glatt H. Human dehydroepiandrosterone sulfotransferase. Purification, molecular cloning, and characterization. Ann N Y Acad Sci. 1995;774:59–72. doi: 10.1111/j.1749-6632.1995.tb17372.x. [DOI] [PubMed] [Google Scholar]
- 21.Her C, Wood TC, Eichler EE, Mohrenweiser HW, Ramagli LS, Siciliano MJ, Weinshilboum RM. Human hydroxysteroid sulfotransferase SULT2B1: two enzymes encoded by a single chromosome 19 gene. Genomics. 1998;53:284–95. doi: 10.1006/geno.1998.5518. [DOI] [PubMed] [Google Scholar]
- 22.Meloche CA, Falany CN. Expression and characterization of the human 3 beta-hydroxysteroid sulfotransferases (SULT2B1a and SULT2B1b) J Steroid Biochem Mol Biol. 2001;77:261–9. doi: 10.1016/s0960-0760(01)00064-4. [DOI] [PubMed] [Google Scholar]
- 23.He D, Meloche CA, Dumas NA, Frost AR, Falany CN. Different subcellular localization of sulphotransferase 2B1b in human placenta and prostate. Biochem J. 2004;379:533–40. doi: 10.1042/BJ20031524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Comer KA, Falany JL, Falany CN. Cloning and expression of human liver dehydroepiandrosterone sulphotransferase. Biochem J. 1993;289:233–40. doi: 10.1042/bj2890233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Falany CN. Human cytosolic sulfotransferases. Properties, physiological functions, and toxicology. In: Lash L, editor. Drug Metabolism and Transport Molecular Methods and Mechanisms. Totowa, NJ: Humana Press; 2005. pp. 341–78. [Google Scholar]
- 26.Vree TB, van den Biggelaar-Martea M, Verwey-van Wissen CP, Vree JB, Guelen PJ. Pharmacokinetics of naproxen, its metabolite O-desmethylnaproxen, and their acyl glucuronides in humans. Biopharm Drug Dispos. 1993;14:491–502. doi: 10.1002/bdd.2510140605. [DOI] [PubMed] [Google Scholar]
- 27.Franssen EJ, Moolenaar F, de Zeeuw D, Meijer DK. Low molecular weight proteins as carriers for renal drug targeting: naproxen coupled to lysozyme via the spacer l-lactic acid. Pharm Res. 1993;10:963–9. doi: 10.1023/a:1018946219057. [DOI] [PubMed] [Google Scholar]
- 28.Rodrigues AD, Kukulka MJ, Roberts EM, Ouellet D, Rodgers TR. [O-methyl 14C]naproxen O-demethylase activity in human liver microsomes: evidence for the involvement of cytochrome P4501a2 P4502c9/10. Drug Metab Dispos. 1996;24:126–36. [PubMed] [Google Scholar]
- 29.Anttila M, Haataja M, Kasanen A. Pharmacokinetics of naproxen in subjects with normal and impaired renal function. Eur J Clin Pharmacol. 1980;18:263–8. doi: 10.1007/BF00563009. [DOI] [PubMed] [Google Scholar]
- 30.Fersht A. A Guide to Enzyme Catalysis and Protein Folding. New York: W.H. Freeman; 1999. Structure, Mechanism in Protein Science. [Google Scholar]
- 31.Barnett AC, Tsvetanov S, Gamage N, Martin JL, Duggleby RG, McManus ME. Active site mutations and substrate inhibition in human sulfotransferase 1A1 and 1A3. J Biol Chem. 2004;279:18799–805. doi: 10.1074/jbc.M312253200. [DOI] [PubMed] [Google Scholar]
- 32.Gamage NU, Duggleby RG, Barnett AC, Tresillian M, Latham CF, Liyou NE, McManus ME, Martin JL. Structure of a human carcinogen-converting enzyme, SULT1A1. Structural kinetic implications substrate inhibition. J Biol Chem. 2003;278:7655–62. doi: 10.1074/jbc.M207246200. [DOI] [PubMed] [Google Scholar]