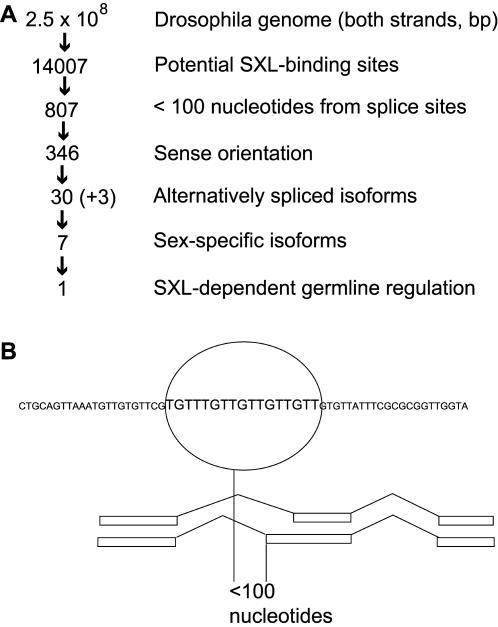
Figure 1. Computational search identifies potential targets of SXL.
(A) Step-wise rationale for the identification of biologically relevant SXL targets. The number of potential candidates with SXL binding sites remaining after each step is indicated. (+3) represents that the three known targets of SXL (Sxl, tra, msl2) were also identified. (B) Schematics of how the search program works. Overlapping nucleotide windows are scanned for sequences that match the consensus-binding site. When a binding site is identified, the search program determines whether it is within 100 nucleotides of a splice site, whether it is in the sense orientation, and whether there is evidence of alternative splicing for that site.