Figure 6. Diagram illustrating the proposed mechanism of DNA relaxation catalyzed by type IA topoisomerases.
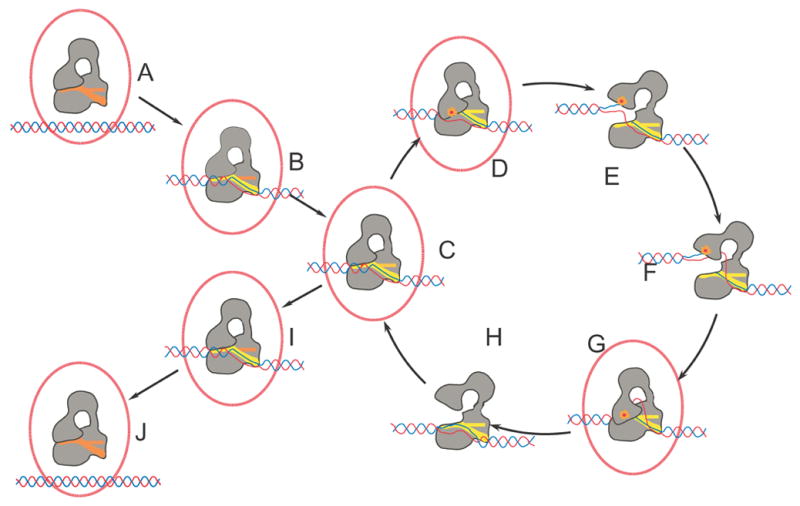
The proposed mechanism involves several transient conformational intermediates both of the protein and the DNA. The sequence of the steps is hypothetical. It is very likely that more states are involved in the cycle. Processivity by the enzyme requires that after one relaxation event the protein continues to another relaxation cycle without releasing the DNA. The selection of states B and I as outside the main cycle is arbitrary. In the diagram, the protein is shown in grey and the DNA in red/blue. The proposed intermediates whose structures are known are surrounded by a red ellipse. The orange dot represents the presence of the covalent protein/DNA complex, which at this stage is hypothetical and has not been observed. The ssDNA binding groove is shown in red or yellow. Helix O, which is found in Domain IV and is part of the binding groove, is shown as a horizontal bar. The color of the binding groove and helix O reflects its conformation. It is red in the closed form, orange in the intermediate form, and yellow in the open form.
Entrance to the cycle
A. Enzyme and supercoiled DNA prior to catalysis. The enzyme is in the closed conformation as found in the apo structure of E. coli DNA topoisomerase III 6.
B. The ssDNA starts invading the ssDNA binding groove and Domain IV in the protein changes conformation to accept the ssDNA. This state is illustrated by the closed form described here.
Processive cycle
C. The ssDNA approaches the active site concomitant with the movement of helix O. The overall structure of the protein resembles the closed form, but the ssDNA and helix O have changed conformation and the scissile phosphate approaches the active site. This state is illustrated by the intermediate form described here.
D. Entrance of the ssDNA into the protein triggers a conformational change to create a catalytically competent active site, as observed in the structure of the complex of topoisomerase III-Y328F with ssDNA 11 and the open form described here.
E. The enzyme opens, bridges the gap between the broken ends of the cleaved DNA, and allows passage of the other strand between the separated ends and into the central hole of the protein. No structures are known of this state, but the structure of a 30 kDa fragment of E. coli topoisomerase I comprising Domains II and III 29 may illustrate the extent of the conformational changes.
F. Enzyme-catalyzed strand passage of the second strand of DNA into the central hole of the enzyme. No structure is known of this intermediate.
G. Following strand passage, the enzyme closes, trapping the passing DNA strand inside the protein. Once the gate is closed, the cleaved strand is religated by the enzyme. The complex of topoisomerase III-Y328F with ssDNA 11 and the open form described here illustrate this state.
H. The cycle is completed by the enzyme opening to release both the religated strand and the one that was passed through the gap. No structure is known of this intermediate.
During the cycle, one DNA strand has passed through the other to change the topology of the DNA and alter the linking number by 1. At this point, the enzyme can continue in the cycle and perform another relaxation event or exit the cycle and release the DNA. The reaction always proceeds toward relaxation of the DNA.
Exit of the cycle
I. The ssDNA starts exiting the ssDNA binding groove and Domain IV in the protein changes conformation to expel the ssDNA. This state is illustrated by the closed form described here.
J. Upon complete release of the DNA, the enzyme returns to the apo conformation 6. The DNA has been relaxed by 1 during each round of the cycle.
