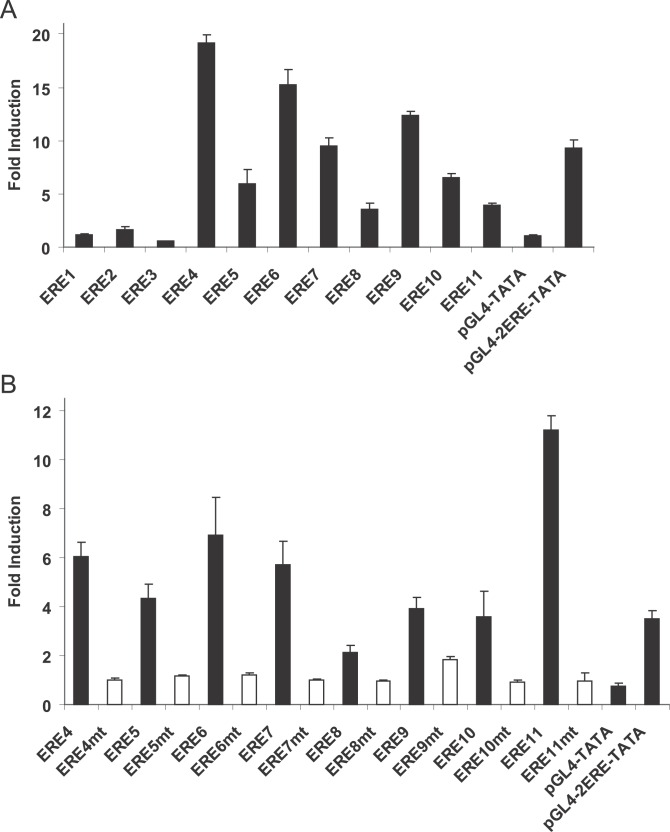
Figure 4. ERE Sequences in ER ChIP-PET Binding Sites Are Functional Transcriptional Enhancers.
(A) ER ChIP-PET binding sites were cloned into the pGL4-TATA luciferase reporter construct and transfected into MCF-7 cells that have been grown in hormone depleted medium for at least three days. The cells were treated with either ethanol or 10 nM estradiol for 18–24 h before harvesting for luciferase activity. pGL4-TATA and pGL4-2ERE-TATA (two copies of the vitellogenin ERE cloned upstream of TATA box of pGL4-TATA) were used as negative and positive controls, respectively. The cells were also cotransfected with the HSV-TK renilla vector as an internal control for transfection efficiency. The data represent the average of three individual experiments ± standard error of mean. The binding site coordinates and their adjacent genes are: ERE1 and ESR1, Chromosome 6: 152029288–152029705; ERE2 and ESR1, Chromosome 6: 152071268–152071889; ERE3 and FOXA1, Chromosome 14: 37189409–37189699; ERE4 and GREB1, Chromosome 2: 11589053–11589737; ERE5 and GREB1, Chromosome 2: 11621762–11622024; ERE6 and GREB1, Chromosome 2: 11622967–11623504; ERE7 and GREB1, Chromosome 2: 11630097–11630780; ERE8 and PGR, Chromosome 11: 100554271–100554807; ERE9 and PGR, Chromosome 11: 100712072–100712428; ERE10 and CA12, Chromosome 15: 61467060–61467460; ERE11 and TFF1, Chromosome 21: 42669273–42670075.
(B) Putative ERE motifs in ER ChIP-PET binding sites were mutated and examined in transient transfection studies as described in (A).