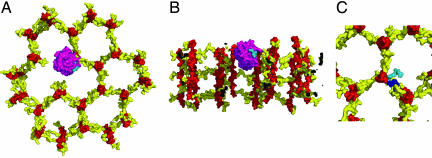
Fig. 4.
Possible interaction of PGRPs with the bacterial cell wall. (A) Top view of a structural model of a PGRP-IβC molecule bound to cell wall PGN. PGRP-IβC and the cell wall are shown in molecular surface representation with the glycan strands of PGN in red, the peptide stems in yellow, PGRP-IβC in purple, and the PGRP-bound peptide stem in cyan. This model was constructed by docking PGRP-IβC onto a GMPP unit of a perpendicular model of the cell wall (17) in which the PGN strands are orthogonal to the cell membrane. The strands form a honeycomb pattern, with pore sizes determined by the extent of cross-linking. Small pores are formed by cross-linking each PGN strand to three neighboring strands. The PGRP-IβC molecule is situated in an incompletely cross-linked region of the growing cell well where a missing PGN strand creates a larger pore. (B) Side view of the model in A, in which the cell wall has been cut away to expose the bound PGRP protein. (C) Comparison of cross-linked and PGRP-bound peptide stems. A cross-linked peptide stem in the model of cell wall PGN is shown in blue. The same peptide stem, but in its PGRP-bound conformation, is shown in cyan.