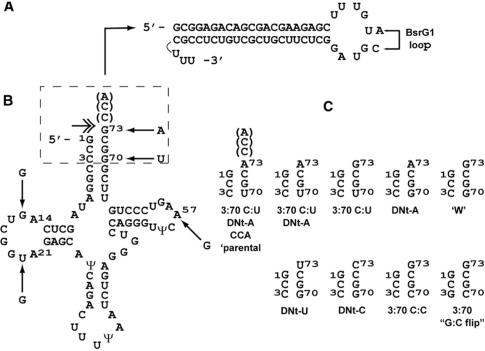
Figure 1.
(A) tat/rev shRNA, as transcribed from the U6 promoter. The hairpin orientation is sense–loop–anti-sense. In some derivatives, additional nucleotides were changed in the loop as shown to create a BsrG1 site for cloning convenience. (B) tRNALys3-tat/rev shRNAs. tRNALys3 wild-type sequence is shown starting with the mature tRNALys3 5′ end; arrows indicate SELEX-derived mutations in the S4tRNALys3 variant; link to base of tat-rev hairpin in (A) is shown schematically. The double headed arrow indicates the location of cleavage required to release the tat/rev hairpin with a sequence identical to that in (A). The dashed box indicates the acceptor stem/DNt region which was varied and shown in (C). (C) Acceptor stem/DNt variants. Only mutated bases relative to wild-type are indicated; all constructs lack the CCA linker between the discriminator nucleotide and the first base of the hairpin unless otherwise indicated. DNt-X indicates a change of the predominant (G) discriminator nucleotide to the indicated base. Third acceptor stem base-pair mutations relative to wild-type are also shown. ‘W’: wild-type acceptor stem. The acceptor stem/DNt nucluotide variants were constructed in both the SELEX4 (St4tRNALys3) and wild-type (+) tRNALys3 backgrounds.