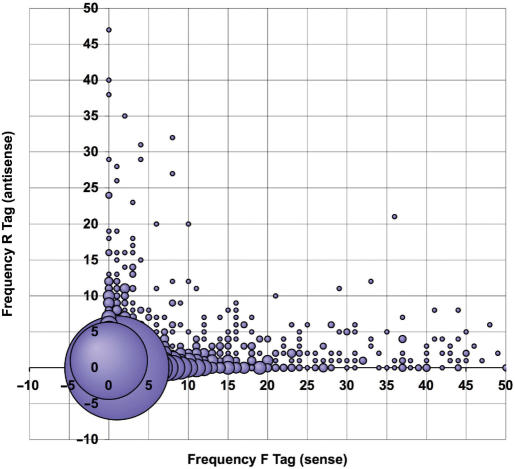
Figure 1.
The abundance of antisense transcripts in Giardia. To assess the abundance and distribution of sense and antisense messages throughout the Giardia genome, all open reading frames (ORFs) and their location with respect to the assembled contigs were downloaded from the genome project database (www.mbl.edu/Giardia), along with trophozoite SAGE tags with respect to the ORFs. A computer algorithm was created to generate a list of all ORFs and the frequency of their forward (sense) and reverse (antisense) SAGE tags for both primary and alternative transcripts. For each ORF, primary and alternative SAGE tags were added, yielding a total forward (sense) and reverse (antisense) tag frequency. The coordinate of each ORF on the graph is depicted by its forward (sense) tag frequency on the x-axis and its reverse (antisense) tag frequency on the y-axis. The size of the bubble at each coordinate is reflective of the number of ORFs with a given forward (sense):reverse (antisense) tag ratio.