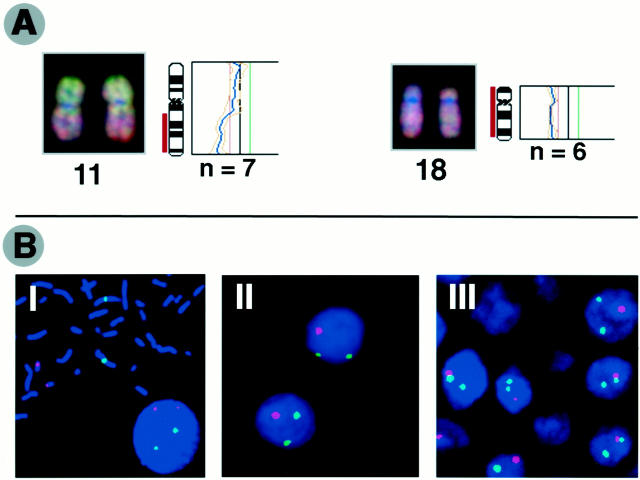
Figure 2.
A: Representative examples of CGH digital images and corresponding profiles illustrating losses of chromosome11q and 18. Tumor DNA was labeled using green dUTP and normal reference DNA with red dUTP. The color ratio values 0.8, 1.2, and 1.5 were used as thresholds for chromosomal losses, gains, and amplifications, respectively. B: Representative pictures of FISH analysis. I: Signals of centromere probes specific for chromosomes 1 (green) and 18 (red) on metaphase from normal human lymphocytes. II: Deletions of chromosome 18 detected on paraffin-embedded section. III: Losses of chromosome 18 on interphase touch preparation. For the FISH analysis on paraffin-embedded sections, the centromere probe of chromosome 18 was labeled using digoxigenin-11-dUTP and then detected using sheep anti-digoxigenin-rhodamine (red). The centromere probe specific for chromosome 1 was directly labeled using spectrum-green dUTP. In the interphase analysis on touch preparations, centromere probes specific for chromosomes 1 and 18 were labeled using spectrum-green dUTP and spectrum-red dUTP, respectively.