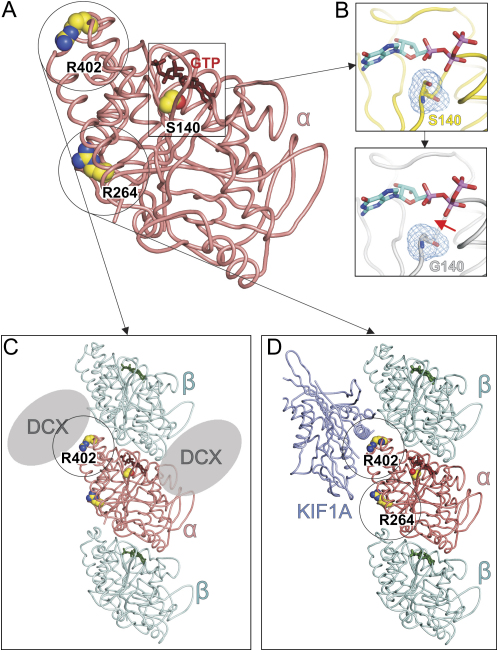
Figure 7.
Mapping of the Mutations onto the α-Tubulin Structure
(A) The structure of α-tubulin (pdb: 1JFF). The three mutations are highlighted as spheres in atomic coloring (nitrogen: blue; oxygen: red; carbon: yellow). The bound GTP molecule is shown as red sticks.
(B) Close-up view of the GTP binding site of α-tubulin. Serine 140, located on the T4 loop, is depicted as a stick presentation in atomic coloring according to (A) with its solvent accessible surface shown as blue wire. The second panel shows a model of the S140G mutation, the red arrow indicating extra space generated by the mutation and potentially responsible for disrupting the interaction with GTP.
(C) Schematic presentation of the tubulin:doublecortin complex based on a structural model from Moores (Moores et al., 2004). The position of arginine 402 (R402), located at the beginning of the H11-H12 loop, is highlighted with a circle (α-tubulin: salmon; β-tubulin: cyan; doublecortin: gray ellipse).
(D) Cartoon presentation of the complex between tubulin and the kinesin KIF1A (pdb: 2HXF). KIF1A is colored in slate. The positions of arginine 264 (R264), located at the surface of the molecule at the loop between H8 and S7, and of argininine 402 (R402), are highlighted by circles.