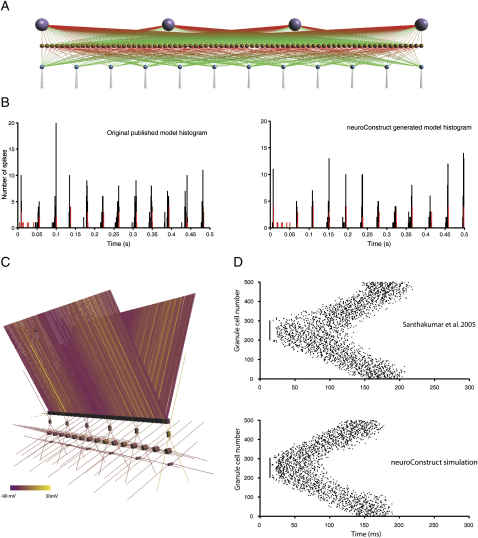
Figure 6.
Implementation of Existing Network Models in neuroConstruct
(A) Visualization of a 1D GrC layer network model from Maex and De Schutter (1998). MFs (bottom) are connected via excitatory synapses to GrCs (middle), which are in turn connected to GoCs (top). The GrCs receive inhibitory connections from GoCs.
(B) Spike time histograms (bin size, 1 ms) as produced by the script files released with the original publication (left) and for the neuroConstruct model of the network (right). Spikes for the GrCs are in black and the GoCs are in red.
(C) Replication of a network model of the dentate gyrus (Santhakumar et al., 2005). The model consists of (from the top down) 500 GrCs with two dendritic branches, 6 basket cells, 15 mossy cells, and 6 hilar cells. The 10,000+ synaptic connections have been removed for clarity. The network receives a brief perforant path focal stimulation, mainly on the central 100 GrCs. Cell coloring reflects network activity 110 ms after stimulation.
(D) Raster plots of dentate gyrus GrC activity in the original published model and in the neuroConstruct implementation of the network.