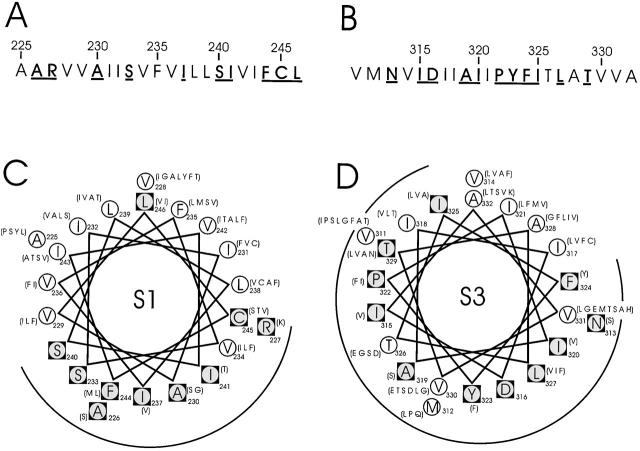
Figure 2.
Sequence characteristics of S1 and S3 segments in Shaker. (A and B) Amino acid sequences of S1 and S3. Boldface underlined residues are those conserved in 120 S1 and 135 S3 K+ channel sequences, using a BLAST search (Altschul et al. 1997) of the nonredundant database. (C and D) α-helical projections of S1 and S3. Conserved residues, inscribed in black squares, are identified by two criteria. First, the most common residue is found in over 65% of instances; second, no more than four alternative substitutes are observed, and these must all be of similar chemical character. Listed in parentheses are alternative amino acids found at equivalent positions in other K+ channels.