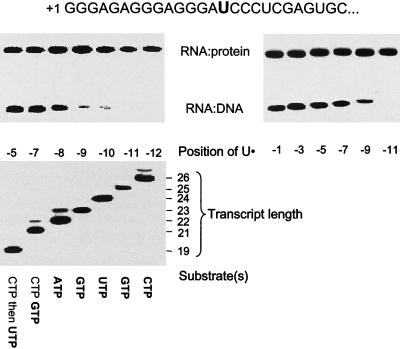
Figure 1.
Crosslinking of RNA to DNA and RNAP. A 120-bp template that directs synthesis of a transcript with the sequence indicated (Top) was constructed by PCR amplification of pPK10 (8). Start up complexes that extended to +15 were formed by incubation of His6-T7 RNAP in the presence of GTP, ATP, and U● (bold). The complexes were immobilized on Ni2+-agarose beads, and the transcripts were incrementally extended by sequential cycles of washing and incubation with the substrates indicated (11). Transcripts were labeled during each cycle by inclusion of the [α-32P]NTP indicated in bold, and each sample was divided into two portions. One portion was examined directly by electrophoresis in 20% gels to verify appropriate extension of the transcript (Lower; the position of the U analog in the transcript is expressed relative to the 3′ end of the RNA at −1). Crosslinking of the transcripts in the other portion was activated by exposure to NaBH4 (13), and the samples were analyzed by PAGE in a 12% gel in the presence of 6 M urea (Upper). Whereas crosslinking to the RNAP was observed at all positions from −5 to −12, efficient crosslinking to the DNA was observed only from −5 to −8. Similar results were obtained using other templates (PK10, PK12, PK13, PK14, D2, and DT3; see ref. 11) that allow U● to be positioned from −1 to −11 (Right). RNAP⋅RNA complexes were identified by their sensitivity to proteinase K and retention on Ni2+ agarose beads; RNA⋅DNA complexes had a mobility that corresponds to the template strand crosslinked to the expected transcript.