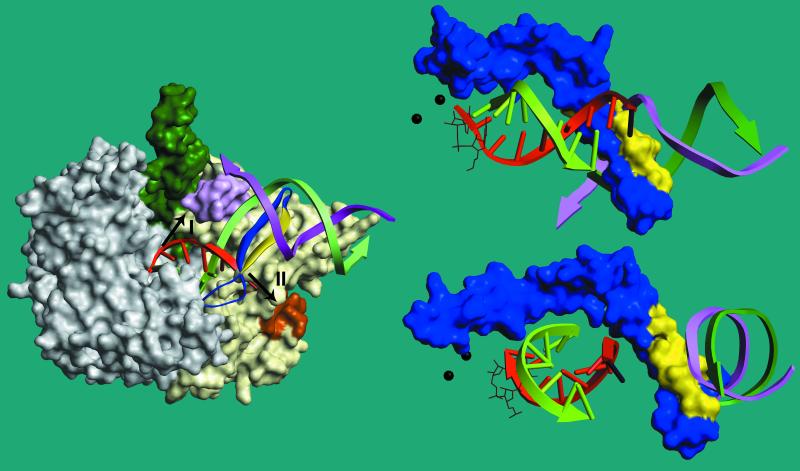
Figure 6.
Model of a T7 RNAP transcription complex. The structures shown are based on the experimentally determined structure of the T7 RNAP initiation complex (6). In the polymerase, the N-terminal domain is tan, the thumb domain is green, the intercalating hairpin that directs the template strand into the active site (residues 230–250) is pink, the N-terminal surface-exposed binding site for exogenous RNA (21) is brown, the specificity loop () is dark blue, and the 7-aa interval to which the −9 crosslink is made (744) is yellow. A putative 8-bp RNA⋅DNA hybrid has been modeled into the DNA binding cleft by homology modeling to the TaqI DNA polymerase primer/template complex (20) by superimposing D537, D812, and Y639 in T7 RNAP (PDB ID QLN), with the corresponding residues in TaqI DNAP (PDB ID TAU) using weblab viewerpro 3.5 (Molecular Simulations, Waltham, MA). The template strand is green, the non-template strand cyan, and the newly made RNA red. In this view, the promoter is to the right, the active site is at the left end of the RNA⋅DNA hybrid, and the front of the cleft obscures the active site and much of the template strand of the RNA⋅DNA hybrid. Arrows show the suggested exit pathways for the nascent transcript as proposed by Cheetham and Steitz (6) or in this work (arrows I and II, respectively). To the right of the overall complex are shown close-up views of the specificity loop, the RNA⋅DNA hybrid, and the binding region of the promoter from two different perspectives. The complex has been rotated such that the view is now under the specificity loop and the template strand of the RNA⋅DNA hybrid is now visible. The first two nucleotides of the nascent RNA are shown in wireframe, and the alpha carbons of the two Asp residues that define the active site (D537, D812) are shown as black spheres. The RNA has been extended by 1 nt (black) to show the proximity of the modeled base at −9 to the specificity loop. In this model, 2–3 nt of unpaired template strand would be required to transit the specificity loop and reestablish duplex DNA at the upstream border of the transcription bubble.