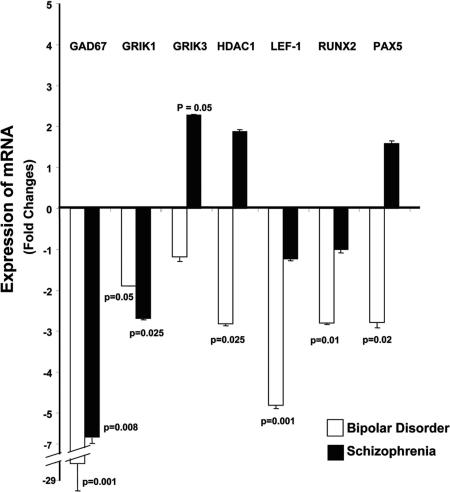
Fig. 2.
QRT-PCR validation of microarray-based GEP data. The results for GAD67, HDAC1, LEF1, Runx2, PAX5, and GRIK1/3 show changes in the same direction as those observed with microarray. The error bars are very small; however, the P value of 0.001 is robust for the REST analysis that was used. The error bars for GAD67 appear to be larger than those of the other genes because of the proportions of the graph needed to accommodate the data for all of the genes. On a percent basis, the error bars for GAD67 when expressed as a percent of the mean are relatively similar to those for the other genes.