Figure 6.
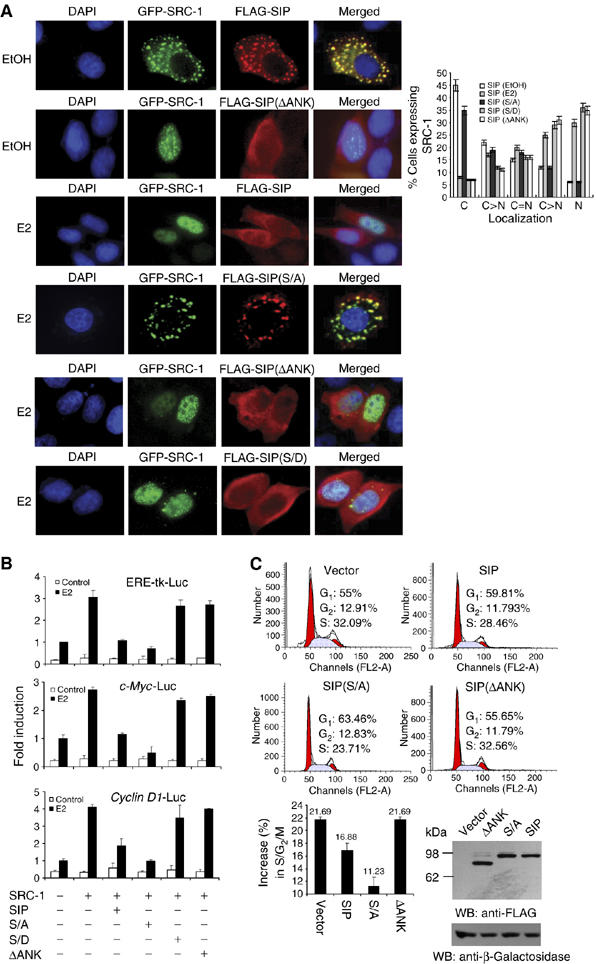
The molecular mechanism involved in sequestration of SRC-1 by SIP. (A) The effect of SIP, SIP(S/A), SIP(ΔANK), and SIP(S/D) on the subcellular localization of SRC-1. pEGFP-SRC-1 and FLAG-tagged SIP, or SIP mutants were cotransfected into MCF-7 cells. Twenty-four hours after the transfection, the cells were left untreated or treated with E2 for 30 min and EGFP fluorescence and rhodamine staining of FLAG were visualized by fluorescence microscopy. Subcellular distribution of SRC-1 was scored in at least 100 cells. The results are means±s.d. of three independent determinations (right panel). (B) The effect of SIP, SIP(S/A), SIP(ΔANK), and SIP(S/D) on ER-regulated gene transcription. MCF-7 cells were cotransfected with the indicated reporter constructs and expression plasmids. After treatment with E2 for 12 h, the cells were collected for the measurement of the reporter activity. Each bar represents the mean±s.d. for triplicate experiments. (C) The effect of SIP, SIP(S/A), and SIP(ΔANK) on E2-stimulated cell cycle progression. MCF-7 cells were transfected with wild-type SIP or SIP mutants and grown in estrogen-depleted media for 2 days. The cells were then treated with E2 for 12 h and the cell cycle profile was analyzed by FCAS. A representative flow cytometry data (upper panels) and the average changes compared with no treatment in the percentage of cells in G0/G1, S, and G2/M phases from three independent experiments (lower panel) are shown. The expression of SIP and its mutants was measured by Western blotting with cotransfection of an E. coli lacZ construct to monitor the transfection efficiency.
