Figure 5.
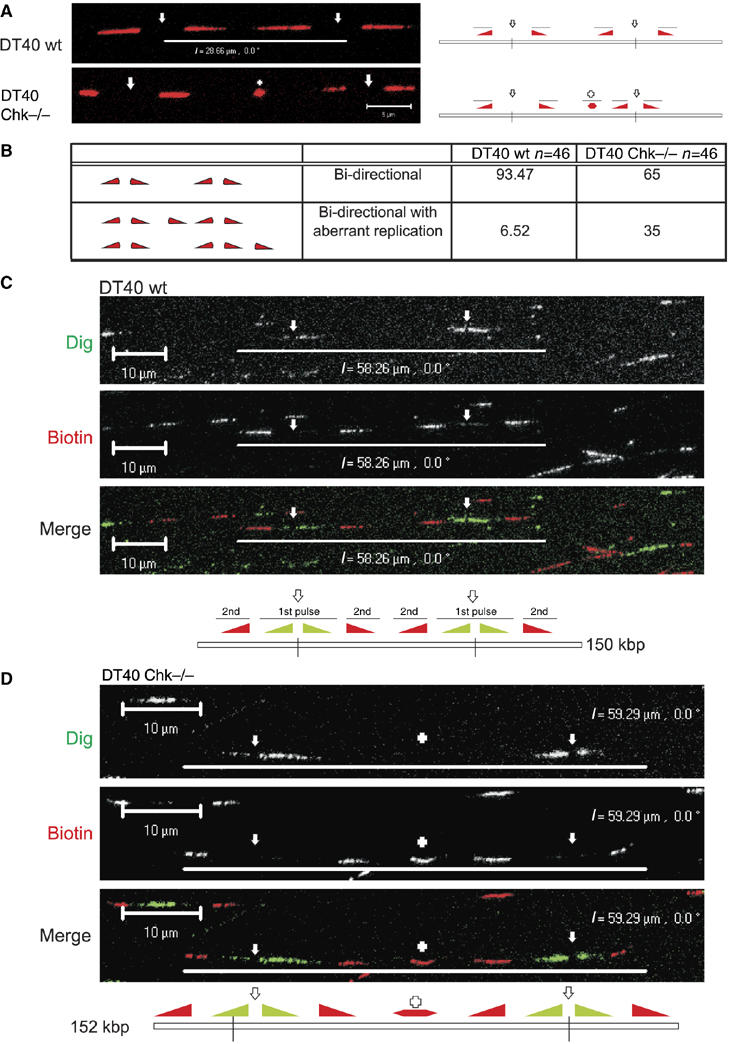
Unprogrammed activation of DNA synthesis in Chk1-deficient DT40 cells. Chk1−/− and isogenically matched wt DT40 cells were labelled with BrdU for 24 h and pulse labelled with biotin-dUTP (30 min; (A, B)) or Dig-dUTP, followed by biotin-11-dUTP (both 30 min; (C, D)). In panel (A) are shown sites of biotin-dU incorporation (red) in single, isolated DNA fibres from wt and Chk1−/− DT40 cells. DNA fibres were labelled with BrdU to ensure that only single fibres were used for the analysis; for simplicity the BrdU labelling is not shown (some examples are provided in Supplementary Figure S4 and S5). Replicon structure was defined by the organisation of the growing forks (diagram (panel A); filled triangles represent the direction of fork growth from central origins, depicted by open arrows; + represents a novel or aberrant replication structure) using fibres with two active replicons (four labelled forks). The relative frequency of normal (bidirectional) and aberrant replication structures (within 25 kb of normal forks) was scored (panel B). Unprogrammed initiation in Chk1−/− cells was confirmed using a combination of Dig-dU and biotin-dU to define replicon structure (panels C, D). DNA fibres were immunolabelled to visualise isolated fibres with two active origins (∼250 kb DNA). Such replicons contain two pairs of growing forks that were labelled first with Dig and then with biotin (panels C, D). In Chk1−/− DT40 (panel D), but not in wt controls (panel C), unprogrammed initiation contributes to synthesis of the unreplicated DNA between the active growing forks—in this example, a new initiation event (+) occurred in the centre of the fibre during the second pulse. Diagrams of the structure of the labelled replicons are shown (panels C, D), using nomenclature described in panel A. Scale bars (5 μm (panel A) and 10 μm (panels C, D)).
