Abstract
The most common sub-variant of papillary thyroid carcinoma (PTC) is the so-called follicular variant (FVPTC), which is a particularly problematic lesion and can be challenging from a diagnostic viewpoint even in resected lesions. Although fine needle aspiration cytology is very useful in the diagnosis of PTC, its accuracy and utility would be greatly facilitated by the development of specific markers for PTC and its common variants. We used the recently developed Applied Biosystems 1700 microarray system to interrogate a series of 11 benign thyroid lesions and conditions and 14 samples of PTC (six with classic morphology and eight with follicular variant morphology). TaqMan® reverse transcriptase-polymerase chain reaction was used to validate the expression portfolios of 50 selected transcripts. Our data corroborates potential biomarkers previously identified in the literature, such as LGALS3, S100A11, LYN, BAX, and cluster of differentiation 44 (CD44). However, we have also identified numerous transcripts never previously implicated in thyroid carcinogenesis, and many of which are not represented on other microarray platforms. Diminished expression of metallothioneins featured strongly among these and suggests a possible role for this family as tumour suppressors in PTC. Fifteen transcripts were significantly associated with FVPTC morphology. Surprisingly, these genes were associated with an extremely narrow repertoire of functions, including the major histocompatibility complex and cathepsin families.
Keywords: Papillary thyroid carcinoma, Biomarker, Follicular variant, Microarray, TaqMan® PCR
Introduction
Papillary thyroid carcinoma (PTC) is the most common endocrine malignancy and encompasses a variety of morphological/architectural variants, all of which are characterized by a distinctive nuclear appearance. In recent years, PTC has become an important paradigm of solid tumour molecular pathogenesis principally arising from intensive investigation prompted by the Chernobyl accident.
The discovery of ret rearrangements [12, 13, 35] and their association with radiation [42] was followed by the demonstration of the BRAF V600E mutation [21, 30] found more commonly associated with sporadic PTC in non-radiation exposed populations [22, 32]. In the past, our group and others have noted an association between classic morphology and the BRAF V600E mutation and between variant morphology and ret rearrangements particularly ret/PTC-3 [11, 39]. Similarly ret/PTC-3 appears to strongly correlate with the solid/follicular variant seen commonly in children exposed to the Chernobyl fallout [42]. Nevertheless, the utility of these genetic lesions to diagnostic pathology and clinical practice has remained negligible.
Recently, gene expression microarray technology has been used to attempt to identify clinically relevant biomarkers of malignancy related to the thyroid [2, 6, 9, 10, 20, 28]. The discovery of such a biomarker or panel of biomarkers allied to the gold standard triage method of fine needle aspiration cytology (FNAC) would represent a significant advancement in the treatment of the solitary thyroid nodule.
An intriguing but commonly occurring variant of PTC is known as follicular variant (FVPTC). This lesion, which by definition retains the classic nuclear features of PTC, shows no evidence of the architectural papillae. FVPTC may be a controversial lesion due to interobserver variation in its pathological diagnosis [25]. Further, the occurrence of follicular patterned lesions with poorly or incompletely developed nuclear features may occur, which are easily dismissed as benign thyroid nodules. This has led to the controversial designation “well-differentiated tumour of uncertain malignant potential” for tumours of this type [45].
It is clear that this is a complex and contentious area, and that further work needs to be done to ascertain the underlying molecular biology of this particular variant. Recently, inroads into elucidation of molecular pathways underpinning PTC have been carried out using microarray studies. The overriding objective of these investigations was to identify clinically useful biomarkers. However, the majority of these studies have analysed PTC as though it were a homogenous singular entity without deference in a detailed manner to sub-variants and, in particular, the most common variant (FVPTC). The identification of specific biomarkers of FVPTC and a deeper understanding of its origins are clearly warranted.
The aim of this expression microarray study using a novel microarray platform was twofold: to identify markers that distinguish PTC from benign thyroid tissue and lesions and, secondly, to identify potential markers and further explore the molecular pathology of FVPTC.
Materials and methods
Patients and tissue samples
Tissue from 25 thyroid resections was collected prospectively from patients undergoing partial or total thyroidectomy for a variety of reasons at St. James’s Hospital, Dublin. The study had approval of the local ethics committee and informed consent was obtained from each patient by the clinical team before surgery. Small samples (<1 cm) were divided and immediately snap-frozen in liquid nitrogen for storage at −80°C until use. Histopathological examination of formalin-fixed paraffin-embedded sections was performed by two pathologists (SF and MT) for diagnostic categorisation. Classification of neoplastic tissue was made according to a recognised system [24]. The cohort comprised 11 benign lesions or conditions including follicular adenoma, nodular goitre, normal thyroid tissue, and Graves’s thyroiditis. The remaining 14 samples were diagnosed as PTC and comprised six classical morphology PTC and eight FVPTC (see Table 1). Immediately before RNA extraction, frozen sections were cut and stained to confirm the presence of representative lesional tissue with morphology corresponding to that noted in the diagnostic formalin-fixed paraffin-embedded sections.
Table 1.
Sample cohort
| Identifier | Diagnosis |
|---|---|
| N1 | Normal thyroid tissue |
| N2 | Normal thyroid tissue |
| N3 | Lymphocytic thyroiditis |
| N4 | Nodular hyperplasia |
| N5 | Follicular adenoma |
| N6 | Nodular hyperplasia with focal lymphocytic thyroiditis |
| N7 | Nodular hyperplasia |
| N8 | Follicular adenoma |
| N9 | Nodular hyperplasia |
| N10 | Follicular adenoma |
| N11 | Grave’s thyroiditis |
| T1 | Solid/FVPTC |
| T2 | FVPTC |
| T3 | PTC classic morphology |
| T4 | FVPTC-oxyphil |
| T5 | FVPTC |
| T6 | FVPTC |
| T7 | PTC classic morphology |
| T8 | FVPTC |
| T9 | PTC classic morphology |
| T10 | FVPTC |
| T11 | PTC classic morphology |
| T12 | FVPTC |
| T13 | PTC classic morphology |
| T14 | PTC classic morphology |
List of the 11 benign and 14 malignant lesions that were used in this study.
FV follicular variant; PTC papillary thyroid carcinoma
RNA isolation and characterization
Samples were ground in liquid nitrogen and homogenised in RLT buffer (Qiagen, UK). RNA was then extracted using the RNeasy Mini Kit with optional on-column RNase-free DNase digestion (Qiagen) according to the manufacturer’s instructions. RNA quantity was determined using UV spectrophotometry. RNA quality was assessed using the RNA 6000 Nano LabChip® Kit in conjunction with the Agilent 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany).
Microarray analysis
Applied Biosystems Human Genome Survey Arrays were used to analyse the transcriptional profiles of thyroid RNA samples in this study. Digoxigenin-UTP-labelled cRNA was generated and linearly amplified from 5 μg of total RNA using Applied Biosystems Chemiluminescent RT-IVT Labelling Kit v 2.0 using manufacturer’s protocol. 10 μg of labelled cRNA were hybridized to each pre-hybed microarray in a 1.5-ml volume at 55°C for 16 h. Array hybridization and chemiluminescence detection were performed using Applied Biosystems Chemiluminescence Detection Kit following manufacturer’s protocol. Images were collected for each microarray using the 1700 analyser. Images were auto-gridded and the chemiluminescent signals were quantified, corrected for background and spot, and spatially normalized.
TaqMan® PCR validation
Sufficient RNA remained from 20 of the initial 25 samples for TaqMan® polymerase chain reaction (PCR) validation in a series of 50 targets. RNA was reverse transcribed using a High Capacity cDNA Archive Kit (Applied Biosystems, CA, USA). Primers and probes for TaqMan® PCR were obtained by using Applied Biosystems’ pre-designed TaqMan® Gene Expression Assays. PCR was carried out using an ABI PRISM 7900 Sequence Detection System (Applied Biosystems). Analysis of relative gene expression data was performed using the ΔΔCT method [23] with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control/reference assay.
Statistical analysis
Microarrays were analysed using Spotfire DecisionSite™ for Functional Genomics (Spotfire AB, Goteborg, Sweden) and R version 1.9.1, a free language and environment for statistical computing and graphics (R Development Core Team, 2004). Arrays were initially normalized, and genes were deemed undetectable and, therefore, excluded from final gene lists if they had a signal-to-noise ratio of less than three (S/N < 3) in greater than 18 of the 25 arrays.
An ANOVA test was used to generate p values for statistical differences between groups. Their p values were then adjusted for multiple comparisons using the technique described by Benjamini and Yekutieli [3]. Genes were deemed statistically different between groups if they had an adjusted p < 0.05 and an average fold-change difference of greater than 2. Hierarchical clustering was performed based on the statistically different genes to determine whether samples grouped appropriately. Gene ontology analysis was performed using an online database known as the Panther classification system (http://www.pantherdb.org). Correlations between microarray and TaqMan® expression data were measured using the Pearson coefficient.
Results
Unsupervised clustering of all 25 samples demonstrated clustering into two major groups (data not shown) comprising the N group and the T group (see Table 1). There was no tendency for FVPTC to cluster independently with classic morphology PTC, confirming the close relationship of these variants of PTC. To identify potential markers of malignancy, ANOVA with false discovery rate correction was used to compare the benign and malignant thyroid cohorts. A p value cut-off of <0.05 and fold-change difference of ≥2 yielded 236 statistically significant probes. Of these, 172 corresponded to fully annotated probes and are listed in Table 2.
Table 2.
Genes differentially expressed in malignant vs benign thyroid tissue
| Gene Name | Gene Symbol | Adjusted p value | 1700 probe ID |
|---|---|---|---|
| Genes up-regulated in malignant vs benign | |||
| Active BCR-related gene | ABR | 0.014482 | 154399 |
| Adaptor-related protein complex 2, alpha 1 subunit | AP2A1 | 0.0312 | 115368 |
| Apoptosis, caspase activation inhibitor | AVEN | 0.030998 | 203738 |
| BCL2-associated X protein | BAX | 0.009782 | 146510 |
| BH3 interacting domain death agonist | BID | 0.021424 | 131216 |
| Brain abundant, membrane attached signal protein 1 | BASP1 | 0.024214 | 198318 |
| Brain acyl-CoA hydrolase | BACH | 0.014566 | 133876 |
| Bromodomain adjacent to zinc finger domain, 1A | BAZ1A | 0.03538 | 209809 |
| Calcium/calmodulin-dependent protein kinase I | CAMK1 | 0.041887 | 157712 |
| Cathepsin S | CTSS | 0.046544 | 105790 |
| CD44 antigen (homing function and Indian blood group system) | CD44 | 0.044181 | 133604 |
| Chemokine (C-X-C motif) ligand 16 | CXCL16 | 0.043234 | 199059 |
| Chromosome 1 open reading frame 38 | C1orf38 | 0.049072 | 202924 |
| CLIP-170-related protein | CLIPR-59 | 0.023629 | 102205 |
| Docking protein 1, 62 kDa (downstream of tyrosine kinase 1) | DOK1 | 0.041898 | 204989 |
| Epidermodysplasia verruciformis 1 | EVER1 | 0.003348 | 175569 |
| FXYD domain containing ion transport regulator 5 | FXYD5 | 0.01444 | 154607 |
| FXYD domain containing ion transport regulator 5 | FXYD5 | 0.023629 | 112771 |
| Galactose-4-epimerase, UDP | GALE | 0.047363 | 141143 |
| Genethonin 1 | GENX-3414 | 0.016836 | 124360 |
| Hypothetical gene BC008967 | BC008967 | 0.015683 | 108526 |
| Hypothetical protein FLJ10849 | FLJ10849 | 0.013822 | 224983 |
| Hypothetical protein FLJ22531 | FLJ22531 | 0.024391 | 145918 |
| Hypothetical protein MGC4607 | MGC4607 | 0.006507 | 211836 |
| Intercellular adhesion molecule 1 (CD54), human rhinovirus receptor | ICAM1 | 0.028746 | 109070 |
| Jun dimerization protein p21SNFT | SNFT | 0.043301 | 144215 |
| Lectin, galactoside-binding, soluble, 3 (galectin 3) | LGALS3 | 0.034491 | 179836 |
| Major vault protein | MVP | 0.0312 | 212354 |
| Matrix metalloproteinase 14 (membrane-inserted) | MMP14 | 0.038682 | 152076 |
| Milk fat globule-EGF factor 8 protein | MFGE8 | 0.02392 | 144588 |
| Mst3 and SOK1-related kinase | MST4 | 0.042028 | 112198 |
| neuronal cell adhesion molecule | NRCAM | 0.011178 | 106462 |
| Phospholipase D3 | PLD3 | 0.034491 | 143388 |
| Promyelocytic leukemia | PML | 0.016018 | 217558 |
| Protein inhibitor of activated STAT protein PIASy | PIASY | 0.00536 | 153434 |
| Protein tyrosine phosphatase, receptor type, E | PTPRE | 0.048653 | 221568 |
| Rho GDP dissociation inhibitor (GDI) beta | ARHGDIB | 0.043853 | 143589 |
| S100 calcium binding protein A11 (calgizzarin) | S100A11 | 0.019933 | 145550 |
| Similar to rat tricarboxylate carrier-like protein | BA108L7.2 | 0.025387 | 179870 |
| SP110 nuclear body protein | SP110 | 0.0312 | 113484 |
| Stimulated by retinoic acid gene 6 | FLJ12541 | 0.043234 | 193986 |
| Syndecan 3 (N-syndecan) | SDC3 | 0.048804 | 143980 |
| Tax interaction protein 1 | TIP-1 | 0.006673 | 119665 |
| TBC1 domain family, member 2 | TBC1D2 | 0.029907 | 205982 |
| Tenascin C (hexabrachion) | TNC | 0.032355 | 143831 |
| Thymosin, beta 4, Y chromosome | TMSB4Y | 0.040937 | 193911 |
| Tissue inhibitor of metalloproteinase 1 (erythroid potentiating activity, collagenase inhibitor) | TIMP1 | 0.032306 | 134692 |
| Topoisomerase (DNA) II alpha 170 kDa | TOP2A | 0.040937 | 135302 |
| Transforming growth factor, beta 1 | TGFB1 | 0.016836 | 170749 |
| Transgelin 2 | TAGLN2 | 0.031971 | 172296 |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta polypeptide | YWHAH | 0.039009 | 188379 |
| v-yes-1 Yamaguchi sarcoma viral related oncogene homolog | LYN | 0.016175 | 194134 |
| Genes down-regulated in malignant vs benign | |||
| Aldehyde oxidase 1 | AOX1 | 0.003227 | 106573 |
| Ankyrin 2, neuronal | ANK2 | 0.035357 | 155780 |
| Aspartate beta-hydroxylase | ASPH | 0.002064 | 221656 |
| Aspartate beta-hydroxylase | ASPH | 0.019666 | 114180 |
| ATPase, Cu++ transporting, beta polypeptide | ATP7B | 0.044511 | 198852 |
| Brain-specific protein p25 alpha | p25 | 0.023629 | 120622 |
| Casein kinase | LOC149420 | 0.022382 | 149347 |
| Cellular retinoic acid binding protein 1 | CRABP1 | 0.008315 | 100295 |
| Centromere protein J | CENPJ | 0.0312 | 164563 |
| Ceroid-lipofuscinosis, neuronal 5 | CLN5 | 0.011021 | 205999 |
| Chloride channel Kb | CLCNKB | 0.040418 | 176266 |
| Chondroitin beta1,4 N-acetylgalactosaminyltransferase | ChGn | 0.013148 | 101140 |
| Chromosome 11 open reading frame 8 | C11orf8 | 0.001977 | 174025 |
| Chromosome 11 open reading frame 8 | C11orf8 | 0.004148 | 108279 |
| Chromosome 21 open reading frame 4 | C21orf4 | 0.0042 | 156895 |
| Clusterin-like 1 (retinal) | CLUL1 | 0.019631 | 186062 |
| Component of oligomeric golgi complex 3 | COG3 | 0.003664 | 129212 |
| Coxsackie virus and adenovirus receptor | CXADR | 0.004648 | 108284 |
| Crystallin, alpha B | CRYAB | 0.030418 | 190274 |
| Cytosolic sialic acid 9-O-acetylesterase homolog | CSE-C | 0.040993 | 213856 |
| Dicarbonyl/l-xylulose reductase | DCXR | 0.001977 | 103350 |
| DnaJ (Hsp40) homolog, subfamily B, member 4 | DNAJB4 | 0.043853 | 103618 |
| ERO1-like beta (S. cerevisiae) | ERO1LB | 0.013962 | 207998 |
| Extracellular link domain containing 1 | XLKD1 | 0.039738 | 195865 |
| Family with sequence similarity 13, member A1 | FAM13A1 | 0.019631 | 116936 |
| Fatty acid binding protein 4, adipocyte | FABP4 | 0.014832 | 150137 |
| Fc fragment of IgG binding protein | FCGBP | 0.001965 | 118361 |
| Fibroblast growth factor receptor 2 | FGFR2 | 0.0042 | 110548 |
| FLJ35740 protein | FLJ35740 | 0.020224 | 101102 |
| Friedreich ataxia region gene X123 | X123 | 0.032602 | 133505 |
| Glutamate-ammonia ligase (glutamine synthase) | GLUL | 0.014649 | 175147 |
| Glycine amidinotransferase (l-arginine/glycine amidinotransferase) | GATM | 0.013962 | 111904 |
| Glycoprotein M6A | GPM6A | 0.011739 | 215326 |
| Growth hormone receptor | GHR | 0.017721 | 190306 |
| HLA complex group 4 | HCG4 | 0.025807 | 191199 |
| Hypothetical protein BC009561 | LOC119710 | 0.003227 | 211319 |
| Hypothetical protein BC019238 | LOC120379 | 0.013438 | 201200 |
| Hypothetical protein FLJ13204 | FLJ13204 | 0.003227 | 145066 |
| Hypothetical protein FLJ13842 | FLJ13842 | 0.016448 | 208504 |
| Hypothetical protein FLJ14054 | FLJ14054 | 0.049072 | 202017 |
| Hypothetical protein FLJ20154 | FLJ20154 | 0.014428 | 143310 |
| Hypothetical protein FLJ20513 | FLJ20513 | 0.019493 | 154130 |
| Hypothetical protein FLJ32110 | FLJ32110 | 0.015507 | 229492 |
| Hypothetical protein FLJ32343 | FLJ32343 | 0.012208 | 116902 |
| Hypothetical protein FLJ33516 | FLJ33516 | 0.03965 | 224600 |
| Hypothetical protein FLJ37549 | FLJ37549 | 0.001956 | 218577 |
| Hypothetical protein FLJ39378 | FLJ39378 | 0.005853 | 163223 |
| Hypothetical protein FLJ40021 | FLJ40021 | 0.023629 | 174198 |
| Hypothetical protein LOC134285 | LOC134285 | 0.018694 | 163671 |
| Hypothetical protein MGC10946 | MGC10946 | 0.022382 | 195982 |
| Hypothetical protein MGC14425 | MGC14425 | 0.015445 | 161569 |
| Hypothetical protein MGC17299 | MGC17299 | 0.026062 | 168452 |
| Hypothetical protein MGC17943 | MGC17943 | 0.0042 | 147296 |
| Hypothetical protein MGC23980 | MGC23980 | 0.018694 | 224619 |
| Hypothetical protein MGC24047 | MGC24047 | 0.001956 | 138122 |
| Hypothetical protein MGC33607 | MGC33607 | 0.033547 | 100645 |
| Ionized calcium binding adapter molecule 2 | IBA2 | 0.0042 | 179489 |
| KIAA0390 gene product | KIAA0390 | 0.014832 | 119936 |
| KIAA0703 gene product | KIAA0703 | 0.032602 | 146652 |
| Lectin, mannose-binding, 1 | LMAN1 | 0.031092 | 179632 |
| Leiomodin 1 (smooth muscle) | LMOD1 | 0.022352 | 120404 |
| Likely ortholog of rat SNF1/AMP-activated protein kinase | SNARK | 0.044605 | 157942 |
| LIM domain kinase 2 | LIMK2 | 0.002409 | 151439 |
| Low density lipoprotein-related protein 1B (deleted in tumors) | LRP1B | 0.00536 | 209464 |
| Low density lipoprotein-related protein 2 | LRP2 | 0.040937 | 114919 |
| Matrilin 2 | MATN2 | 0.0042 | 167316 |
| Metallothionein 1A (functional) | MT1A | 0.013822 | 204773 |
| Metallothionein 1A (functional)|metallothionein 1E (functional)|metallothionein 1K| metallothionein 2A | MT1A|MT2A|MT1E|MT1K | 0.027037 | 146368 |
| Metallothionein 1A (functional)|metallothionein 1E (functional)|metallothionein 2A| metallothionein 1K | MT1A|MT2A|MT1K|MT1E | 0.043841 | 182305 |
| Metallothionein 1A (functional)|metallothionein 2A|metallothionein 1K| metallothionein 1E (functional) | MT1A|MT1K|MT1E|MT2A | 0.011739 | 223856 |
| Metallothionein 1B (functional) | MT1B | 0.019631 | 174119 |
| Metallothionein 1F (functional) | MT1F | 0.024726 | 144569 |
| Metallothionein 1G | MT1G | 0.0192 | 164525 |
| Metallothionein 1G | MT1G | 0.03965 | 171539 |
| Metallothionein 1J | MT1J | 0.008315 | 227956 |
| Metallothionein 1X | MT1X | 0.008335 | 119685 |
| Metallothionein 1X | MT1X | 0.010748 | 173072 |
| Metallothionein IV | MT4 | 0.007447 | 223241 |
| Methionine adenosyltransferase II, alpha | MAT2A | 0.014428 | 158350 |
| Mitogen-activated protein kinase 4 | MAPK4 | 0.042306 | 131252 |
| Myc-induced nuclear antigen, 53 kDa | MINA53 | 0.011959 | 130284 |
| NIMA (never in mitosis gene a)- related kinase 11 | NEK11 | 0.001965 | 194628 |
| Otospiralin | LOC150677 | 0.018694 | 182360 |
| PDZ/coiled-coil domain binding partner for the rho-family GTPase TC10 | PIST | 0.013822 | 103651 |
| Phospholipase A2 receptor 1, 180 kDa | PLA2R1 | 0.004029 | 134379 |
| Phospholipase C-like 1 | PLCL1 | 0.022657 | 206894 |
| Phosphotidylinositol transfer protein, beta | PITPNB | 0.014428 | 122698 |
| Polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 | PKHD1L1 | 0.0042 | 199896 |
| Polymerase (DNA directed) iota | POLI | 0.003227 | 167492 |
| Potassium channel, subfamily K, member 9 | KCNK9 | 0.000849 | 108648 |
| Potassium channel-interacting protein 4 | KCNIP4 | 0.011447 | 147058 |
| Potassium inwardly-rectifying channel, subfamily J, member 13 | KCNJ13 | 0.008972 | 124187 |
| pp21 Homolog | LOC51186 | 0.004326 | 127636 |
| Pre-B cell leukemia transcription factor 4 | PBX4 | 0.030311 | 199118 |
| Protein kinase, cAMP-dependent, catalytic, beta | PRKACB | 0.023863 | 198878 |
| Protein phosphatase 4, regulatory subunit 2|hypothetical protein LOC151987 | PPP4R2|LOC151987 | 0.011338 | 200919 |
| RAB23, member RAS oncogene family | RAB23 | 0.000659 | 122394 |
| Ras association (RalGDS/AF-6) domain family 6 | RASSF6 | 0.048804 | 119072 |
| Sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | SGCD | 0.011178 | 120415 |
| Serum deprivation response (phosphatidylserine binding protein) | SDPR | 0.011021 | 156433 |
| SH3 and multiple ankyrin repeat domains 2 | SHANK2 | 0.043996 | 193906 |
| Solute carrier family 26, member 7 | SLC26A7 | 0.0042 | 225067 |
| Solute carrier family 26, member 7 | SLC26A7 | 0.005853 | 213530 |
| Solute carrier family 5 (iodide transporter), member 8 | SLC5A8 | 0.031284 | 231731 |
| SPARC related modular calcium binding 2 | SMOC2 | 0.021505 | 135930 |
| Syndecan 2 (heparan sulfate proteoglycan 1, cell surface-associated, fibroglycan) | SDC2 | 0.001258 | 209676 |
| Syntaxin 12 | STX12 | 0.01117 | 199949 |
| T-box 22 | TBX22 | 0.024297 | 177517 |
| Thioredoxin-like, 32 kDa | TXNL | 0.001102 | 192552 |
| Thyroid stimulating hormone receptor | TSHR | 0.02176 | 108606 |
| Tissue inhibitor of metalloproteinase 4 | TIMP4 | 0.023629 | 184795 |
| Trefoil factor 3 (intestinal) | TFF3 | 0.004648 | 114445 |
| Trefoil factor 3 (intestinal) | TFF3 | 0.014428 | 100949 |
| UDP-N-acetyl-alpha-d-galactosamine/polypeptide N-acetylgalactosaminyltransferase 9 (GalNAc-T9) | GALNT9 | 0.031284 | 161042 |
| WEE1 homolog (S. pombe) | WEE1 | 0.024101 | 123533 |
| WW domain containing oxidoreductase | WWOX | 0.012208 | 224298 |
| WW domain containing oxidoreductase | WWOX | 0.024101 | 135080 |
| Zinc finger protein 258 | ZNF258 | 0.013962 | 225961 |
| Zinc finger protein 36, C3H type-like 2 | ZFP36L2 | 0.018837 | 210469 |
Two-tail ANOVA with p value correction yielded 173 probes (52 up-, 121 down-regulated in PTC) significantly different (p < 0.05) between the malignant and benign thyroid tissues.
Supervised hierarchical clustering was performed on the 25 arrays based on the 236 statistically significant probes to determine whether the samples would segregate appropriately. The resulting heat map can be observed in Fig. 1. Benign lesions and tumours clustered together with the exception of one tumour sample that clustered with the benign group (T5).
Fig. 1.
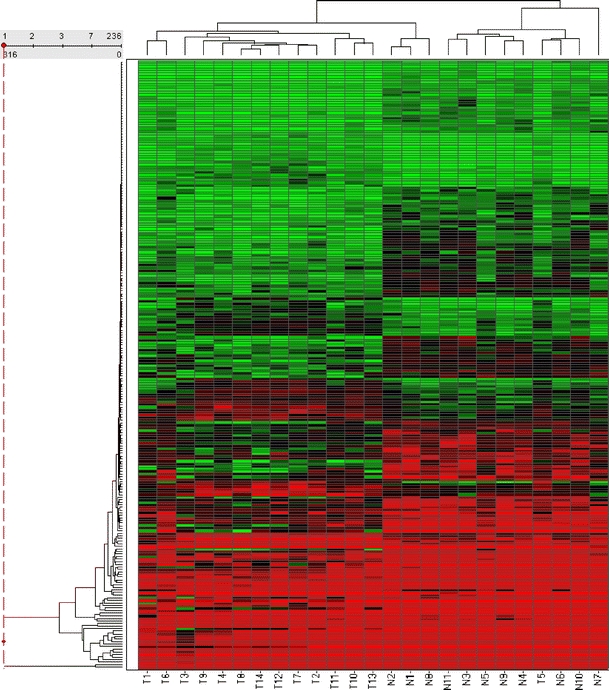
Hierarchical clustering of samples. This heat map shows the clustering of the 25 samples based on the 236 probes found to be differentially regulated in benign vs malignant thyroid tissue. Clustering was performed using the unweighted pair group method with arithmetic mean, with Euclidian distance as the similarity measure. Average value was used as the ordering function
A binomial statistics tool was used to compare classifications of multiple clusters of lists to a reference list (i.e. the complete human genome) to statistically determine over- or under-representation of Panther classification categories. Biological processes over-represented in the up-regulated PTC cohort included tumour suppressor, oncogenesis, DNA replication, cell cycle, and cell adhesion (p < 0.0001). Genes involved in homeostasis and other homeostasis activities were highly over-represented in the down-regulated cohort (p < 0.000001).
ANOVA tests were used to determine which genes were differentially regulated in the FVPTC cohort only. Fifteen genes were identified, including cluster of differentiation 14 (CD14), CD74, CTSC, CTSH, CTSS, DPP6, ETHE1, human leucocyte antigen A (HLA-A), HLA-DMA, HLA-DPB1, HLA-DQB1, HLA-DRA, osteoclast stimulating factor 1 (OSTF1), TDO2, and a previously uncharacterized gene (noname).
Microarray results were validated using a reverse transcription reaction followed by TaqMan® PCR for 50 gene targets. The ΔΔCT method [23] was used to analyse relative gene expression data. GAPDH was used as an endogenous control, and T12 was chosen as an arbitrary calibrator sample. Gene expression profiles for TaqMan® PCR were plotted in conjunction with those for microarray results in Fig. 2. Pearson co-efficient was used to directly compare data from microarray analysis and TaqMan® RT-PCR. Table 3 depicts genes differentially expressed in both benign vs malignant and FVPTC vs classic morphology PTC.
Fig. 2.
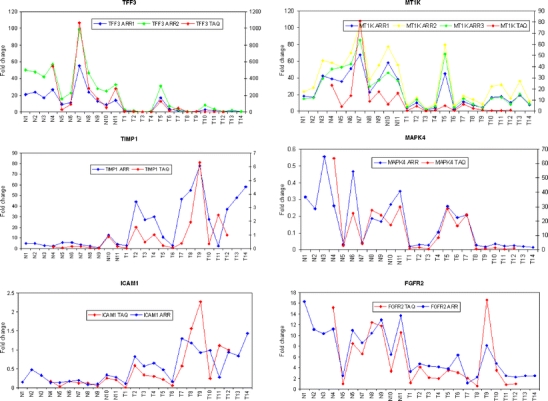
TaqMan® PCR validation of microarray experiments. Profile charts of gene expression levels comparing results obtained by microarray analysis (n = 25) to TaqMan® PCR analysis (n = 20) for six genes. Plots for those genes with multiple probes are also displayed where appropriate
Table 3.
Correlation between TaqMan® and microarray data
| Gene | Pearson’s r coefficient | two-tailed p |
|---|---|---|
| Genes differentially expressed in FVPTC vs classic PTC | ||
| CD14 | 0.83 | <0.0001 |
| CD74a | 0.87 | <0.0001 |
| 0.74 | 0.0002 | |
| CTSC | 0.53 | 0.0170 |
| CTSH | 0.71 | 0.0005 |
| CTSS | 0.62 | 0.0037 |
| DPP6 | 0.76 | 0.0001 |
| ETHE1 | 0.30 | 0.2008 |
| HLA-A | 0.77 | <0.0001 |
| HLA-DMA | 0.75 | 0.0001 |
| HLA-DPB1 | 0.96 | <0.0001 |
| HLA-DQB1a | 0.70 | 0.0006 |
| 0.63 | 0.0031 | |
| HLA-DRA | 0.82 | <0.0001 |
| NONAME | −0.02 | 0.9323 |
| OSTF1 | 0.15 | 0.5171 |
| TDO2 | 0.68 | 0.0009 |
| Genes differentially expressed in benign vs malignant | ||
| BAX | 0.20 | 0.3997 |
| CAMK1 | 0.17 | 0.4771 |
| CD44 | 0.57 | 0.0094 |
| CTSS | 0.62 | 0.0037 |
| CXADR | 0.36 | 0.1152 |
| FGFR2 | 0.84 | <0.0001 |
| GALE | 0.54 | 0.0138 |
| ICAM1 | 0.62 | 0.0038 |
| LYN | 0.45 | 0.0483 |
| MAPK4 | 0.76 | <0.0001 |
| MMP14 | 0.36 | 0.1175 |
| MT1F | 0.92 | <0.0001 |
| MT1Ka | 0.68 | 0.0011 |
| 0.69 | 0.0007 | |
| 0.70 | 0.0006 | |
| MT1Xa | 0.88 | <0.0001 |
| 0.88 | <0.0001 | |
| RAB23 | 0.70 | 0.0006 |
| S100A11 | 0.61 | 0.0041 |
| SDC2 | 0.34 | 0.1415 |
| SDC3 | 0.57 | 0.0087 |
| TFF3a | 0.97 | <0.0001 |
| 0.96 | <0.0001 | |
| TGFB1 | 0.56 | 0.0098 |
| TIMP1 | 0.73 | 0.0003 |
| TIMP4 | 0.80 | <0.0001 |
| TOP2A | 0.07 | 0.7769 |
| TSHR | 0.18 | 0.4440 |
Gene expression profiles for TaqMan® PCR and microarray results were compared using the Pearson coefficient.
aGenes have more than one probe ID on microarray
Discussion
The primary aim of this study was to generate an overview of molecular markers of malignancy in PTC with a view to identifying discriminators between common sub-types (classic PTC and FVPTC), using genome-wide expression microarray technology validated by TaqMan® RT-PCR. To this end, lesions that were well characterized histologically were selected.
The application of microarray analysis designed to identify transcripts strongly associated with each group of interest yielded a gene list of 173 genes that were differentially expressed between cohorts. Significant down-regulation of Coxsackie virus receptor was recorded in the malignant cohort of thyroid lesions. The Coxsackievirus B and adenovirus receptor (CAR) plays a dual role as a homotypic junctional adhesion protein and as a viral receptor. It is biologically plausible that altered expression may impact on the morphology peculiar to PTC given its association with cellular adhesion. CAR has been shown to be differentially expressed in various human adenocarcinomas, and differential expression may represent a new factor in thyroid tumourigenesisigenesis [27].
Rab 23 expression was also down regulated in the malignant cohort. The Rab small G protein family is composed of approximately 40 members. Many of them are ubiquitous and are expressed and participate in transport processes, such as endocytosis and exocytosis [26]. Other gene targets demonstrating significant down-regulation in the malignant group were syndecans 2 and 3. Syndecans are transmembrane proteoglycans expressed on adherent cells. Changes in syndecan expression have been postulated to influence cell adhesion, migration, and the structure of focal contacts and the cytoskeleton [8].
The abundance of metallothionein genes in the cohort of genes down-regulated in PTC is interesting partly because of the sheer number of isoforms detected (Table 2). There have been many studies showing increased metallothionein expression in a plethora of cancer types but few showing decreased expression [4, 7, 33]. Although previous microarray experiments showed metallothionein genes to be down-regulated in thyroid tumours to a certain extent [2, 9, 16], none have detected so many as the current study.
Apart from microarray experiments, there has been little reported in the literature regarding metallothioneins in thyroid cancer. An early report by Nartey et al. [31] showed metallothioneins to be expressed actively in certain human thyroid neoplastic tissues but not in normal thyroid tissue, which would seem to contradict the current authors’ findings. In contrast to this, a later immunohistochemical study showed an absence of metallothionein expression in 13 of 20 PTCs [36]. Interestingly, in three of the seven positive PTCs, metallothionein positivity was restricted to areas of follicular differentiation. In one of the only recent studies, Huang et al. [17] followed up their initial microarray experiment by showing that MT1G is down regulated in PTC via hypermethylation. The biological significance of low metallothionein expression in thyroid tumours is therefore still poorly understood; however, it is interesting to speculate that metallothioneins may have roles as tumour suppressors in thyroid carcinoma.
Many genes identified, such as LGALS3 [9, 16, 20], LYN [46], TFF3 [2, 9, 16], CRABP1 [9, 16], BAX [2], MAPK4 [28], CD44 [16], TIMP1 [20], FGFR2 [9], and S100A11 [20, 40], have been previously reported in both microarray and conventional experiments in thyroid cancer. The data generated in this study corroborates the importance of several biological processes in the progression of thyroid neoplasia. For example, S100A11 expression was up regulated in the PTC cohort compared with benign lesions, paralleling the increased protein expression of this gene target identified at the protein level using immunohistochemistry [29]. S100A11 has also been suggested as a biomarker of malignancy in the context of colorectal carcinoma as long ago as 1995 [41].
Correlation of highlighted features with the current state of knowledge of the molecular pathology of thyroid neoplasia goes some way towards providing an external validation of the data obtained from the AB1700 system. However, additional validation using TaqMan® RT-PCR was performed.
In general, microarray and TaqMan® data correlated well with approx. 80% of comparisons having p < 0.05. Candidate genes were selected contingent on results identified as over-represented biological processes (oncogenesis, cell cycling, DNA replication, and homeostasis) using the Panther binomial statistics tool as opposed to the more traditional method of selecting the most highly disregulated genes. This may account for the poor correlation observed with certain genes. Some genes, such as TFF3 and FGFR2, had excellent correlation between microarray and TaqMan® results, whereas others, such as BAX and TSHR, showed poor correlation despite previous studies implicating them in thyroid cancer [15, 38]. Where there was discordance in the data, it could be accounted for by differences in the sequences targeted by the TaqMan® target sequences and the microarray probe. In those cases, the array and pre-designed TaqMan assays interrogated different exons or alternative splice variants. This finding highlights the importance of matching the targets to be validated from microarray data sets.
Analysis of differentially expressed transcripts in the FVPTC revealed many transcripts showing similar expression level patterns in both FVPTC and classic morphology PTC. Examples of these transcripts are included in Table 2. In addition, somewhat surprisingly, unsupervised clustering of all samples showed no tendency for FVPTC to cluster independently of classic morphology PTC, emphasizing the very close relationship of these PTC variants (data not shown). However, analysis of differentially expressed genes in FVPTC exclusive of those identified in classic morphology PTC revealed 15 genes exhibiting differential expression in FVPTC compared with benign lesions and conditions outwith of classic morphology PTC. These genes displayed a narrow gamut of function represented by the transcripts involved (Table 3). TaqMan® RT-PCR was performed for all of these targets to confirm the array findings and correlation with the array data was strong (see Table 3).
Aberrant expression of two major groups of transcripts was noted in FVPTC. Relatively increased expression of class 1 major histocompatibility complex (MHC) genes (HLA-A) and aberrant expression of class 2 MHC genes (HLA-DMA, HLA-DPB-1, HLA-DQB-1, HLA DRA) and associated genes (e.g. CD74 represents the invariant membrane bound moiety of class II HLA molecules and regulates the biology and functions of MHC class II molecules and CD14 is a surface marker of monocytes/macrophages) were the most significant findings. Additionally, relatively up-regulated expression of members of the cathepsin family (cathepsin C, cathepsin H, cathepsin S, and TDO2) was striking in the FVPTC group. HLA expression is generally associated with immune functions such as T cell interaction and antigen presentation. The presence of prominent HLA transcript expression, especially among class 2 in FVPTC, is intriguing. One potential cause of this was that tumour-infiltrating leucocytes were responsible for this finding. However, haematoxylin–eosin (H&E)-stained slides of each case were reviewed to specifically identify the degree of tumour infiltration by leucocytes. Although a mild lymphocytic infiltrate was noted in some cases, there was no apparent over representation of lymphocytes in the follicular variant compared to benign lesions and classic morphology PTC. This raises the clear possibility that the findings represent aberrant increased expression of class 2 HLA transcripts by the epithelium of FVPTC. This is an unexpected finding, as over-expression of MHC class 2 molecules would be expected to increase tumour immunogenicity. A similar aberrant expression of HLA transcripts has been recently described in ovarian neoplasms [34]. Rangel et al. [34] concluded over-expression of HLA-DRA might represent a novel biomarker for malignancy, and this also seems biologically plausible in the FVPTC setting. A recent paper has also described HLA-DRA expression in ret/PTC-activated papillary thyroid carcinoma but not in surrounding normal thyroid follicles [19]. Yu et al. [47] showed discordant expression of (CD74Ii) and HLA-DR in Hashimoto thyroiditis, an autoimmune condition associated with increased incidence of PTC and sharing molecular features such as ret/PTC expression [18, 37]. Hwang et al. [19] have drawn attention to aberrant expression of HLA-DRA in ret/PTC-activated PTC. Expression of HLA-DRA may in some way explain the propensity for PTC to metastasize to lymph nodes and often apparently reside there without markedly worsening prognosis.
Cathepsins C (dipeptidyl-peptidase I), H, and S showed up-regulation in FVPTC compared to benign lesions. Cathepsins are a family of proteases that play an important role in protein degradation. They are key players in the proliferative, invasive, and metastatic potential of malignant tumour cells. Their expression in the relatively biologically indolent FVPTC is intriguing, and it remains possible that cathepsins have cellular roles outside of those involved in invasion and dissemination of tumour cells as indeed has been suggested by others [43]. For example, cathepsin L has recently been shown to play a role in nuclear transcriptional activation, and cathepsins are now recognized to play a role in MHC class II antigen presentation [44]. OSTF1 has no defined role in carcinogenesis, although outside of its role in ossification, it is also known to have roles in signal transduction and protein binding, which may be relevant to carcinogenesis and, particularly, FVPTC. A recent paper highlights the role of bone mineralization proteins including osteopontin and osteoclast stimulating factors as potential biomarkers of malignant tumours in general [1]. Indeed, in addition to elevated OSTF1 expression, increased expression of osteopontin was seen in PTC (data not shown); however, in our data, osteopontin does not appear to be specifically up-regulated in FVPTC, and this finding has also been noted by other researchers [14]. In any case, osteopontin is known to be a downstream effector of ret/PTC [5] and mutated BRAF [14], where it acts in association with CD44, another transcript showing increased expression in both classic morphology PTC and FVPTC.
A particular focus of this study was to compare transcriptome profiles for PTCs with classic morphology and FVPTC given the propensity for FVPTC lesions to prove problematic from a diagnostic perspective. Although the study confirms the close relationship between the two most common variants of PTC, a narrow portfolio of genes and, in particular, gene functions was elucidated in the FVPTC cohort. The targets identified are easily amenable to analysis by more established techniques such as TaqMan® RT-PCR, with associated potential as additional markers for application in the FNAC setting. Clearly, the potential biomarkers identified in this study will require prospective evaluation in the context of real clinical diagnostic situations in the future to consolidate their merit as adjunctive tests in the diagnostic setting and to validate their altered expression states in the pathobiology of PTC development.
Acknowledgement
Dr. Smyth and Dr. Finn contributed equally to this study.
References
- 1.Amatschek S, Koenig U, Auer H, Steinlein P, Pacher M, Gruenfelder A, Dekan G, Vogl S, Kubista E, Heider KH, Stratowa C, Schreiber M, Sommergruber W (2004) Tissue-wide expression profiling using cDNA subtraction and microarrays to identify tumor-specific genes. Cancer Res 264:844–856 [DOI] [PubMed]
- 2.Barden CB, Shister KW, Zhu B, Guiter G, Greenblatt DY, Zeiger MA, Fahey TJ 3rd (2003) Classification of follicular thyroid tumors by molecular signature: results of gene profiling. Clin Cancer Res 9:1792–1800 [PubMed]
- 3.Benjamini Y, Yekutieli D (2001) The control of the false discovery rate in multiple testing under dependency. Ann Stat 29:1165–1188 [DOI]
- 4.Cai L, Wang GJ, Xu ZL, Deng DX, Chakrabarti S, Cherian MG (1998) Metallothionein and apoptosis in primary human hepatocellular carcinoma (HCC) from northern China. Anticancer Res 18:4667–4672 [PubMed]
- 5.Castellone MD, Celetti A, Guarino V, Cirafici AM, Basolo F, Giannini R, Medico E, Kruhoffer M, Orntoft TF, Curcio F, Fusco A, Melillo RM, Santoro M (2004) Autocrine stimulation by osteopontin plays a pivotal role in the expression of the mitogenic and invasive phenotype of RET/PTC-transformed thyroid cells. Oncogene 23:2188–2196 [DOI] [PubMed]
- 6.Cerutti JM, Delcelo R, Amadei MJ, Nakabashi C, Maciel RM, Peterson B, Shoemaker J, Riggins GJ (2004) A preoperative diagnostic test that distinguishes benign from malignant thyroid carcinoma based on gene expression. J Clin Invest 113:1234–1242 [DOI] [PMC free article] [PubMed]
- 7.Deng DX, Chakrabarti S, Waalkes MP, Cherian MG (1998) Metallothionein and apoptosis in primary human hepatocellular carcinoma and metastatic adenocarcinoma. Histopathology 32:340–347 [DOI] [PubMed]
- 8.Ding K, Lopez-Burks M, Sanchez-Duran JA, Korc M, Lander AD (2005) Growth factor-induced shedding of syndecan-1 confers glypican-1 dependence on mitogenic responses of cancer cells. J Cell Biol 171:729–738 [DOI] [PMC free article] [PubMed]
- 9.Finley DJ, Arora N, Zhu B, Gallagher L, Fahey TJ 3rd (2004) Molecular profiling distinguishes papillary carcinoma from benign thyroid nodules. J Clin Endocrinol Metab 89:3214–3223 [DOI] [PubMed]
- 10.Finley DJ, Zhu B, Barden CB, Fahey TJ 3rd (2004) Discrimination of benign and malignant thyroid nodules by molecular profiling. Ann Surg 240:425–436 [DOI] [PMC free article] [PubMed]
- 11.Finn SP, Smyth P, O’Leary J, Sweeney EC, Sheils O (2003) Ret/PTC chimeric transcripts in an Irish cohort of sporadic papillary thyroid carcinoma. J Clin Endocrinol Metab 88:938–941 [DOI] [PubMed]
- 12.Fusco A, Grieco M, Santoro M, Berlingieri MT, Pilotti S, Pierotti MA, Della Porta G, Vecchio G (1987) A new oncogene in human thyroid papillary carcinomas and their lymph-nodal metastases. Nature 328:170–172 [DOI] [PubMed]
- 13.Grieco M, Santoro M, Berlingieri MT, Melillo RM, Donghi R, Bongarzone I, Pierotti MA, Della Porta G, Fusco A, Vecchio G (1990) PTC is a novel rearranged form of the ret proto-oncogene and is frequently detected in vivo in human thyroid papillary carcinomas. Cell 60:557–563 [DOI] [PubMed]
- 14.Guarino V, Faviana P, Salvatore G, Castellone MD, Cirafici AM, De Falco V, Celetti A, Giannini R, Basolo F, Melillo RM, Santoro M (2005) Osteopontin is overexpressed in human papillary thyroid carcinomas and enhances thyroid carcinoma cell invasiveness. J Clin Endocrinol Metab 90:5270–5278 [DOI] [PubMed]
- 15.Hermann S, Sturm I, Mrozek A, Klosterhalfen B, Hauptmann S, Dorken B, Daniel PT (2001) Bax expression in benign and malignant thyroid tumours: dysregulation of wild-type P53 is associated with a high Bax and P21 expression in thyroid carcinoma. Int J Cancer 92:805–811 [DOI] [PubMed]
- 16.Huang Y, Prasad M, Lemon WJ, Hampel H, Wright FA, Kornacker K, LiVolsi V, Frankel W, Kloos RT, Eng C, Pellegata NS, de la Chapelle A (2001) Gene expression in papillary thyroid carcinoma reveals highly consistent profiles. Proc Natl Acad Sci USA 98:15044–15049 [DOI] [PMC free article] [PubMed]
- 17.Huang Y, de la Chapelle A, Pellegata NS (2003) Hypermethylation, but not LOH, is associated with the low expression of MT1G and CRABP1 in papillary thyroid carcinoma. Int J Cancer 104:735–744 [DOI] [PubMed]
- 18.Hunt JL, Baloch ZW, Barnes L, Swalsky PA, Trusky CL, Sesatomi E, Finkelstein S, LiVolsi VA (2002) Loss of heterozygosity mutations of tumor suppressor genes in cytologically atypical areas in chronic lymphocytic thyroiditis. Endocr Pathol 13:321–330 [DOI] [PubMed]
- 19.Hwang ES, Kim DW, Hwang JH, Jung HS, Suh JM, Park YJ, Chung HK, Song JH, Park KC, Park SH, Yun HJ, Kim JM, Shong M (2004) Regulation of signal transducer and activator of transcription 1 (STAT1) and STAT1-dependent genes by RET/PTC (rearranged in transformation/papillary thyroid carcinoma) oncogenic tyrosine kinases. Mol Endocrinol 18:2672–2684 [DOI] [PubMed]
- 20.Jarzab B, Wiench M, Fujarewicz K, Simek K, Jarzab M, Oczko-Wojciechowska M, Wloch J, Czarniecka A, Chmielik E, Lange D, Pawlaczek A, Szpak S, Gubala E, Swierniak A (2005) Gene expression profile of papillary thyroid cancer: sources of variability and diagnostic implications. Cancer Res 65:1587–1597 [DOI] [PubMed]
- 21.Kimura ET, Nikiforova MN, Zhu Z, Knauf JA, Nikiforov YE, Fagin JA (2003) High prevalence of BRAF mutations in thyroid cancer: genetic evidence for constitutive activation of the RET/PTC–RAS–BRAF signaling pathway in papillary thyroid carcinoma. Cancer Res 63:1454–1457 [PubMed]
- 22.Lima J, Trovisco V, Soares P, Maximo V, Magalhaes J, Salvatore G, Santoro M, Bogdanova T, Tronko M, Abrosimov A, Jeremiah S, Thomas G, Williams D, Sobrinho-Simoes M (2004) BRAF mutations are not a major event in post-Chernobyl childhood thyroid carcinomas. J Clin Endocrinol Metab 89:4267–4271 [DOI] [PubMed]
- 23.Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods 25:402–408 [DOI] [PubMed]
- 24.Lloyd R, DeLellis R, Heitz P, Eng C (eds) (2004) Pathology and genetics of tumours of endocrine Organs. IARC Press
- 25.Lloyd RV, Erickson LA, Casey MB, Lam KY, Lohse CM, Asa SL, Chan JK, DeLellis RA, Harach HR, Kakudo K, LiVolsi VA, Rosai J, Sebo TJ, Sobrinho-Simoes M, Wenig BM, Lae ME (2004) Observer variation in the diagnosis of follicular variant of papillary thyroid carcinoma. Am J Surg Pathol 28:1336–1340 [DOI] [PubMed]
- 26.Marcos I, Borrego S, Antinolo G (2003) Molecular cloning and characterization of human RAB23, a member of the group of Rab GTPases. Int J Mol Med 12:983–987 [PubMed]
- 27.Marsee DK, Vadysirisack DD, Morrison CD, Prasad ML, Eng C, Duh QY, Rauen KA, Kloos RT, Jhiang SM (2005) Variable expression of Coxsackie–adenovirus receptor in thyroid tumors: implications for adenoviral gene therapy. Thyroid 15:977–987 [DOI] [PubMed]
- 28.Mazzanti C, Zeiger MA, Costouros NG, Umbricht C, Westra WH, Smith D, Somervell H, Bevilacqua G, Alexander HR, Libutti SK (2004) Using gene expression profiling to differentiate benign versus malignant thyroid tumors. Cancer Res 64:2898–2903 [DOI] [PubMed]
- 29.Mitselou A, Vougiouklakis TG, Peschos D, Dallas P, Boumba VA, Agnantis NJ (2002) Immunohistochemical study of the expression of S-100 protein, epithelial membrane antigen, cytokeratin and carcinoembryonic antigen in thyroid lesions. Anticancer Res 22:1777–1780 [PubMed]
- 30.Namba H, Nakashima M, Hayashi T, Hayashida N, Maeda S, Rogounovitch TI, Ohtsuru A, Saenko VA, Kanematsu T, Yamashita S (2003) Clinical implication of hot spot BRAF mutation, V599E, in papillary thyroid cancers. J Clin Endocrinol Metab 88:4393–4397 [DOI] [PubMed]
- 31.Nartey N, Cherian MG, Banerjee D (1987) Immunohistochemical localization of metallothionein in human thyroid tumors. Am J Pathol 129:177–182 [PMC free article] [PubMed]
- 32.Nikiforova MN, Ciampi R, Salvatore G, Santoro M, Gandhi M, Knauf JA, Thomas GA, Jeremiah S, Bogdanova TI, Tronko MD, Fagin JA, Nikiforov YE (2004) Low prevalence of BRAF mutations in radiation-induced thyroid tumors in contrast to sporadic papillary carcinomas. Cancer Lett 209:1–6 [DOI] [PubMed]
- 33.Ofner D, Maier H, Riedmann B, Bammer T, Rumer A, Winde G, Bocker W, Jasani B, Schmid KW (1994) Immunohistochemical metallothionein expression in colorectal adenocarcinoma: correlation with tumour stage and patient survival. Virchows Arch 425:491–497 [DOI] [PubMed]
- 34.Rangel LB, Agarwal R, Sherman-Baust CA, Mello-Coelho V, Pizer ES, Ji H, Taub DD, Morin PJ (2004) Anomalous expression of the HLA-DR alpha and beta chains in ovarian and other cancers. Cancer Biol Ther 3:1021–1027 [DOI] [PubMed]
- 35.Santoro M, Carlomagno F, Hay ID, Herrmann MA, Grieco M, Melillo R, Pierotti MA, Bongarzone I, Della Porta G, Berger N, Peix JL, Paulin C, Fabien N, Vecchio G, Jenkins RB, Fusco A (1992) Ret oncogene activation in human thyroid neoplasms is restricted to the papillary cancer subtype. J Clin Invest 89:1517–1522 [DOI] [PMC free article] [PubMed]
- 36.Schmid KW, Greeff M, Hittmair A, Totsch M, Ofner D, Dockhorn-Dworniczak B, Bocker W, Jasani B (1994) Metallothionein expression in normal, hyperplastic, and neoplastic thyroid follicular and parafollicular C cells using monoclonal antimetallothionein antibody E9. Endocr Pathol 5:114–122 [DOI] [PubMed]
- 37.Sheils OM, O’Leary JJ, Uhlmann V, Lattich K, Sweeney EC (2000) ret/PTC-1 activation in Hashimoto thyroiditis. Int J Surg Pathol 8:185–189 [DOI] [PubMed]
- 38.Sheils OM, Sweeney EC (1999) TSH receptor status of thyroid neoplasms-TaqMan RT-PCR analysis of archival material. J Pathol 188:87–92 [DOI] [PubMed]
- 39.Smyth P, Finn S, Cahill S, O’Regan E, Flavin R, O’Leary JJ, Sheils O (2005) ret/PTC and BRAF act as distinct molecular, time-dependant triggers in a sporadic Irish cohort of papillary thyroid carcinoma. Int J Surg Pathol 13:1–8 [DOI] [PubMed]
- 40.Stolf BS, Abreu CM, Mahler-Araujo MB, Dellamano M, Martins WK, de Carvalho MB, Curado MP, Diaz JP, Fabri A, Brentani H, Carvalho AF, Soares FA, Kowalski LP, Hirata R Jr, Reis LF (2005) Expression profile of malignant and non-malignant diseases of the thyroid gland reveals altered expression of a common set of genes in goiter and papillary carcinomas. Cancer Lett 227:59–73 [DOI] [PubMed]
- 41.Tanaka M, Adzuma K, Iwami M, Yoshimoto K, Monden Y, Itakura M (1995) Human calgizzarin; one colorectal cancer-related gene selected by a large scale random cDNA sequencing and Northern blot analysis. Cancer Lett 89:195–200 [DOI] [PubMed]
- 42.Thomas GA, Bunnell H, Cook HA, Williams ED, Nerovnya A, Cherstvoy ED, Tronko ND, Bogdanova TI, Chiappetta G, Viglietto G, Pentimalli F, Salvatore G, Fusco A, Santoro M, Vecchio G (1999) High prevalence of RET/PTC rearrangements in Ukrainian and Belarussian post-Chernobyl thyroid papillary carcinomas: a strong correlation between RET/PTC3 and the solid-follicular variant. J Clin Endocrinol Metab 84:4232–4238 [DOI] [PubMed]
- 43.Turk B, Turk D, Turk V (2000) Lysosomal cysteine proteases: more than scavengers. Biochim Biophys Acta 1477:98–111 [DOI] [PubMed]
- 44.Villadangos JA, Bryant RA, Deussing J, Driessen C, Lennon-Dumenil AM, Riese RJ, Roth W, Saftig P, Shi GP, Chapman HA, Peters C, Ploegh HL (1999) Proteases involved in MHC class II antigen presentation. Immunol Rev 172:109–120 [DOI] [PubMed]
- 45.Williams ED (2000) Two proposals regarding the terminology of thyroid tumors. Int J Surg Pathol 8:181–183 [DOI] [PubMed]
- 46.Yano Y, Uematsu N, Yashiro T, Hara H, Ueno E, Miwa M, Tsujimoto G, Aiyoshi Y, Uchida K (2004) Gene expression profiling identifies platelet-derived growth factor as a diagnostic molecular marker for papillary thyroid carcinoma. Clin Cancer Res 10:2035–2043 [DOI] [PubMed]
- 47.Yu M, Xu M, Savas L, Khan A (1998) Discordant expression of Ii and HLA-DR in thyrocytes: a possible pathogenetic factor in Hashimoto’s thyroiditis. Endocr Pathol 9:201–208 [DOI] [PubMed]


