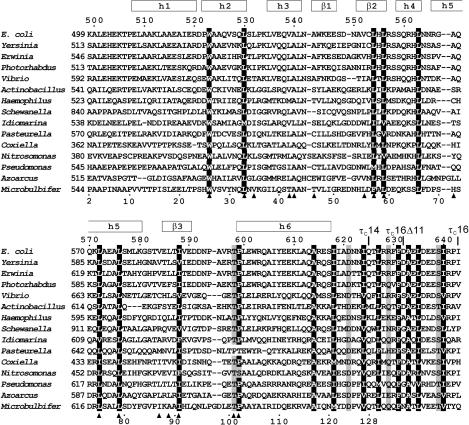
Figure 1.
Sequence alignment of τC16 with τ proteins from different bacteria. The sequence of τC16 comprises the 145 C-terminal residues of the τ subunit of E. coli DNA Pol III holoenzyme (residues 499–643 which constitute Domain V). Its sequence numbering is shown at the top. The constructs τC14, τC16Δ11 and τC16 share the same N-terminus. Their C-termini are indicated above the sequence. The sequence numbering shown below the alignment is that of the NMR sample of τC14 included for reference with previous work (41). The regular secondary structure elements of τC14 determined in this work are shown at the top. Positions with conserved hydrophobic residues are indicated by white characters on a black background. Conserved hydrophilic residues are shaded gray. Filled triangles below the alignment identify residues with side chains buried in the τC14 structure (<5% solvent accessibility). The following sequences from τ proteins are shown (abbreviation, organism, GenBank number): E. coli, Escherichia coli, 43320; Yersinia, Yersinia pseudotuberculosis, 51595342; Erwinia, Erwinia carotovora subsp. Atroseptica, 49610641; Photorhabdus, Photorhabdus lummines subsp. Laumondii tto1, 37527701; Vibrio, Vibrio vulnificus cmcp6, 27361492; Actinobacillus, Actinobacillus pleuropneumoniae serovar 1 STR, 32035419; Haemophilus, Haemophilus somnus, 32029123; Shewanella, Shewanella oneidensis MR-1, 24373576; Idiomarina, Idiomarina loihiensis L2TR, 56460949; Pasteurella, Pasteurella multocida subsp. multocida str. Pm70, 12720609; Coxiella, Coxiella burnetii RSA 493, 29541263; Nitrosomonsa, Nitrosomonas europaea ATCC 19718, 30138336; Pseudomonas, Pseudomonas fluorescens PfO-1, 48731928; Azoarcus, Azoarcus sp. EbN1, 56476074; Microbulbifer, Microbulbifer degradans 2-40, 48862900.