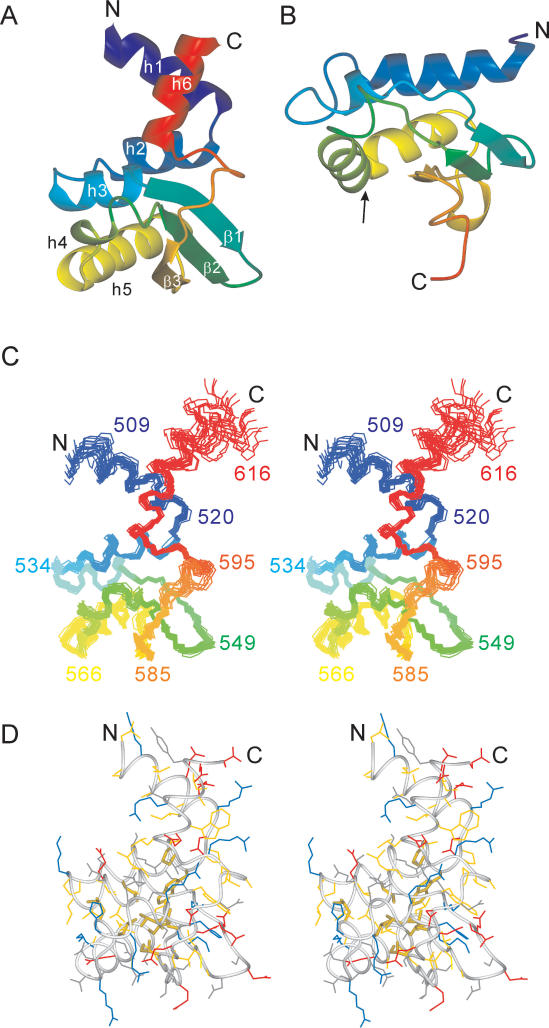
Figure 3.
Solution structure of τC14 and comparison with the KH domain of protein MPN156. (A) Ribbon representation of τC14, including residues 509–609. The secondary structure elements are labeled as in Figure 1. (B) Ribbon representation of MPN156 from Mycoplasma pneumoniae (PDB code: 1PA4). The arrow identifies the RNA-binding helix-loop-helix motif of the KH-domain type II fold. (C) Stereo view of a superposition of the backbone atoms of 20 NMR conformers, including residues 507–617. (D) Stereo view of a heavy-atom representation of the conformer closest to the mean structure of τC14 (only residues 509–609 are shown). The side-chains are color coded in blue (Lys, Arg, His), red (Asp, Glu), yellow (Ala, Cys, Ile, Leu, Met, Phe, Pro, Trp, Val) and gray (Asn, Gln, Ser, Thr, Tyr). The side chains of the strictly conserved hydrophobic residues Trp523, Leu530, Leu554, Leu556, Leu563, Leu572, Leu576, Ile588 and Pro599 are highlighted with broader lines. This figure was prepared using the program MOLMOL (35).