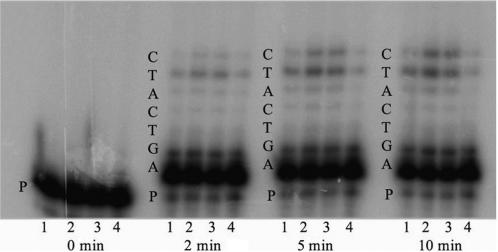
Figure 6.
Example of the in vitro five base-pair missing base primer-extension fidelity assay. Reaction products formed during the gap-filling assay when dCTP is missing from the nucleotide pool are shown. Purified enzymes were incubated with gapped DNA substrate that was radiolabeled at the 5′ end of the primer (Table 2) and a pool of three dNTPs missing dCTP for 2, 5 and 10 min at 37°C as described in the Materials and methods section. The products resulting from incorporation of nucleotides into the primer were resolved on a denaturing acrylamide gel and visualized on a Phosphorimager. Lane 1 is wild type, lane 2 is lambda-loop, lane 3 is 5-Ala loop, lane 4 is 4-Ala loop and P is the primer strand. The length of incubation time is listed underneath each group. The sequence of the template is listed on the side of each group.