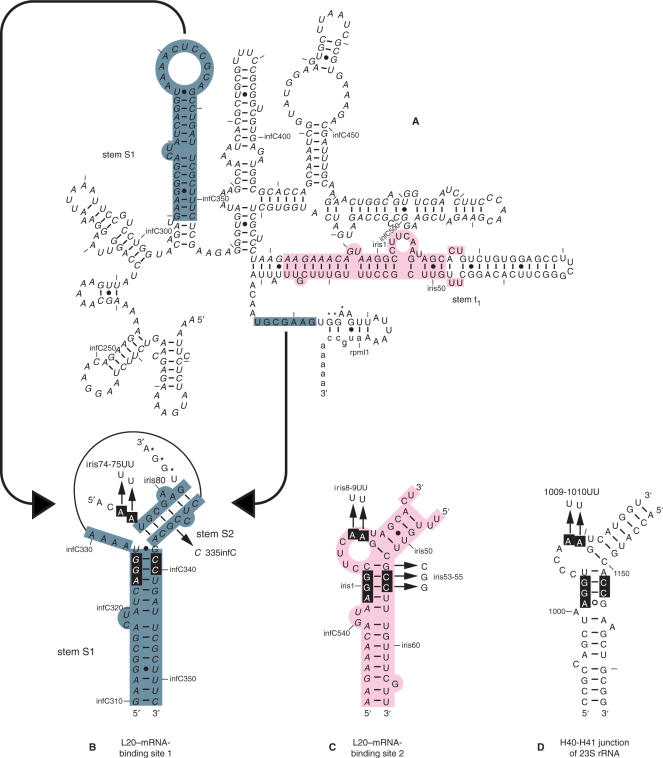
Figure 1.
Secondary structure of the mRNA leader region of the rpmI-rplT operon. (A) Model of the secondary structure of the rpmI translational operator (13). infC, iris (an acronym for infC-rpmI intergenic sequence) and rpmI sequences are italicized, uppercased and lowercased, respectively. Residues are numbered according to infC, iris and rpmI numbering. Every 10th residue is marked with a tick mark and every 50th nucleotide is numbered. Some of the relevant features of the operator (stems S1, S2 and t1 containing the t1 transcriptional terminator) are also indicated. The nucleotides forming the rpmI Shine–Dalgarno sequence are indicated by asterisks. (B) Schematic view of the pseudoknotted site 1. infC and iris sequences and the nucleotides forming the rpmI Shine–Dalgarno sequence are indicated as in (A). The long-range base-pairing interaction which holds the pseudoknot is schematized by two converging arrows starting from the separated interacting partners in (A). (C) Schematic view of site 2. infC and iris sequences are indicated as in (A). (D) Schematic view of the H40-H41 junction of E. coli 23S rRNA. The names of the mutations used in the fluorescent measurement experiments are indicated by arrows pointing to the substituting nucleotides in the schematic representations of the three L20–RNA-binding sites.